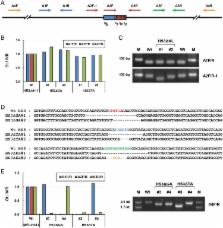
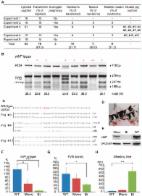
Dear Editor, The pig is an important livestock for food supply and an ideal model for various human diseases. Efficient and precise genetic engineering in pigs holds great promise in agriculture and biomedicine 1 . Using currently available approach, generating specific gene modifications in pigs requires two steps. First, site-specific nucleases such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are used to generate targeted mutations in pig somatic cells. Then the engineered somatic nucleus is used to generate cloned animals using somatic cell nuclear transfer (SCNT) technology 2,3 . The complex design and generation of ZFNs and TALENs, as well as the technical challenges of SCNT, greatly limit the application of this method. Recently a new type of site-specific nuclease was identified in CRISPR (clustered regularly interspaced short palindromic repeat)/Cas (CRISPR-associated) RNA-based adaptive immune system in bacteria and archaea 4 . The Cas9 endonuclease from Streptococcus pyogenes type II CRISPR/Cas system can be engineered to produce targeted genome modification under the guidance of a synthetic single guide RNA (sgRNA) with simple base-pair complementarities with a target genomic DNA sequence 5,6,7 . Although highly efficient one-step generation of mice and rats carrying single or multiple mutations have been achieved through direct injection of CRISPR/Cas system into one-cell embryo 8,9 , it remains to be tested whether the same strategy could be applied to generation of genome-engineered large animals. Here we report the efficient generation of biallelic knockout pigs in one step by direct cytoplasmic injection of Cas9 mRNA and sgRNA into zygotes. To demonstrate the application of genome engineering in disease modeling and agriculture, we chose to target vWF gene, whose deficiency in human causes severe von Willebrand disease (vWD) 10 (Supplementary information, Figure S1). As mouse model does not fully recapitulate the disease phenotype and response to treatment in human patients, a large animal model with similar size of organ and circulatory system is highly desirable. In food industry, bleeding is an important procedure before meat production, as the leftover blood is an ideal medium for bacterial growth. vWF knockout could improve bleeding efficiency for slaughtering procedures and blood collection. We designed an sgRNA targeting exon 5 of the pig vWF gene, which lies in the first trysin-inhibitor-like domain and its mutation could lead to the loss of function of vWF protein. The in vitro developmental efficiencies of embryos injected with Cas9 mRNA/sgRNA (∼79%) and embryos injected with water (∼77%) were both very high and comparable with each other, suggesting that the microinjection manipulation and the Cas9 mRNA/sgRNA had little effect on pig early embryonic development (Supplementary information, Table S1). We next transferred the injected embryos into surrogate pigs to produce piglets. A total of 76 injected embryos were transplanted into 5 surrogate mothers in 5 independent experiments. Three surrogates were successfully pregnant and delivered a total of 16 piglets (Figure 1A). T7 endonuclease I (T7EI) assay showed that 10 piglets contained insertions or deletions (indels) in the targeting site, indicating that vWF mutation efficiency reached about 63% (10/16) (Figure 1A and 1B). We further analyzed the genotype by Sanger sequencing of the PCR products containing the targeting site of each individual piglet. The sequencing results confirmed the indels and revealed that one mutant pig yielding uncut PCR product in the T7EI assay (fourth lane in Figure 1B) contained only one type of mutant allele. Most pigs contained more than two different alleles (Figure 1C, pig #4 and #11), suggesting that the Cas9-mediated DNA cleavage may occur at embryonic stage later than the one-cell stage, as previously observed with ZFN, TALEN and CRISPR-mediated targeting 11 . In each pig, if no wild-type allele was detected by sequencing, we categorized the pig as biallelic mutant. If both wild-type and mutant alleles were observed in the sequence reads, we categorized the pig as monoallelic mutant, although the ratio of wild-type to mutant alleles might not always be 1. In total, we identified six vWF biallelic mutant piglets and five monoallelic mutant piglets (11/16, 68%) (Figure 1A and 1C). Of these piglets, two died shortly after birth and the other 14 survived until now (Figure 1D). The high birth rate (16/76, 21%) and survival rate (14/16, 88%) indicated that Cas9 mRNA/sgRNA had little toxicity to pig embryonic development (Figure 1A). These results demonstrate that the CRISPR/Cas system could efficiently generate both monoallelic and biallelic mutant pigs in one step through direct zygote injection. We further examined whether the vWF mutations cause the vWD phenotypes. Western blot analysis showed that the vWF expression in lung was largely reduced in monoallelic mutant pigs, and was undetectable in biallelic mutant pigs, confirming the vWF knockout at protein levels (Figure 1E). We next performed hematological analysis with pigs having different genotypes. The plasma vWF antigen level was significantly reduced in the monoallelic mutant pig group, and was nearly undetectable in biallelic mutant group (Figure 1F). In human vWD, the level of coagulation factor FVIII may be reduced due to rapid degradation in the absence of vWF 10 . Consistently, the activated partial thromboplastin time (APTT)-based assay for FVIII activity showed a significantly lower activity in the biallelic mutant group, indicating that FVIII level was decreased in the plasma of vWF-null pigs (Figure 1G). No significant differences were observed between wild-type and knockout pigs with regard to other parameters in the hematological analysis (Supplementary information, Table S2). The most prominent phenotype resulting from vWF mutation is a severe bleeding tendency 10 . We found that the bleeding time of biallelic mutant pigs (512 ± 121 min) was much longer than that of wild-type pigs (34 ± 11 min) (Figure 1H). Taken together, our results demonstrate that the one-step generated biallelic mutant pigs developed similar phenotypes of the human vWD. A certain level of off-target cleavage by CRISPR/Cas system was suggested by recent studies in human cell lines 12 , indicating that Cas9-mediated DNA cleavage tolerates small numbers of mismatches between sgRNA and target DNA especially in protospacer adjacent motif (PAM)-distal region 12 . We thus sought to test the possible off-target effects in genome-modified pigs derived from zygote injection. By screening the genome based on sequence homology (> 14 base pair identity to 3′ end of the sgRNA spacer), around 20 potential off-target sites were found (Supplementary information, Figure S2). In all 11 mutant animals tested, none of them contained indels at 2 randomly chosen sites based on T7EI assay (Supplementary information, Figure S2). We further analyzed the potential off-target effects by Sanger sequencing of 10 randomly selected potential off-target sites in 3 biallelic mutant pigs (#9, #10 and #11). None of the sequencing reads had mutations, suggesting no off-target effects at these sites (Supplementary information, Figure S2). However, we could not exclude the possible existence of certain off-target effects due to the high number of potential off-target sites. Thus a systematic off-target evaluation would be needed in the future before the wide application of such knockout pig models. Also, modified CRISPR/Cas-methods with less off-target tendency can be used for genome engineering in pigs and other large animals 13 . Germline transmission of the mutations to next generation is essential for expanding the knockout pig population. To examine the germline transmission potential of the mutations, we dissected an ovary from a monoallelic mutant female pig and a testis from a biallelic mutant male pig by biopsy and analyzed their genotypes by Sanger sequencing. Both the ovary and the testis were morphologically normal (Supplementary information, Figure S3). More than half sequence reads of the ovary and all the sequence reads of the testis were mutant, indicating that the mutations had been efficiently transmitted into the germline (Supplementary information, Figure S3). Considering the highly efficient germline transmission in rodents using the same procedure 14 , germline transmission of the mutations to next generation is highly expected. In summary, we have shown that zygote injection of the CRISPR/Cas system can efficiently generate genome-modified pigs in one step, resulting in rapid establishment of large animal model for important human diseases. The high efficiency of Cas9-mediated genome editing in pigs demonstrated here, together with the versatile functionality of the CRISPR/Cas system such as multiplexed genome editing and epigenetic modification 5 , opens up unlimited possibilities of genome engineering in livestock for applications in agriculture and biomedicine.