- Record: found
- Abstract: found
- Article: found
Rhizosphere suppression hinders antibiotic resistance gene (ARG) spread under bacterial invasion.
Read this article at
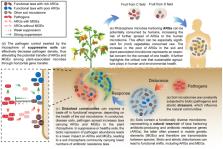
Abstract
The rhizosphere is an extremely important component of the “one health” scenario by linking the soil microbiome and plants, in which the potential enrichment of antibiotic resistance genes (ARGs) might ultimately flow into the human food chain. Despite the increased occurrence of soil-borne diseases, which can lead to increased use of pesticides and antibiotic-producing biocontrol agents, the understanding of the dynamics of ARG spread in the rhizosphere is largely overlooked. Here, tomato seedlings grown in soils conducive and suppressive to the pathogen Ralstonia solanacearum were selected as a model to investigate ARG spread in the rhizosphere with and without pathogen invasion. Metagenomics data revealed that R. solanacearum invasion increased the density of ARGs and mobile genetic elements (MGEs). Although we found ARGs originating from human pathogenic bacteria in both soils, the enrichment was alleviated in the suppressive soil. In summary, the suppressive soil hindered ARG spread through pathogen suppression and had a lower number of taxa carrying antibiotic resistance.
Highlights
-
•
Bacterial pathogen invasion increased the density of ARGs and MGEs within rhizosphere.
-
•
Enriched ARGs were originated from the Invading pathogen and human pathogenic bacteria.
-
•
Suppressive rhizosphere soil could alleviate the ARGs enrichment.
-
•
Lower number of invaded and native taxa carrying ARGs lead to the alleviation.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Call of the wild: antibiotic resistance genes in natural environments.
- Record: found
- Abstract: found
- Article: not found
Antibiotic resistance is ancient.
- Record: found
- Abstract: found
- Article: not found