- Record: found
- Abstract: found
- Article: found
Selected Clostridia Strains from The Human Microbiota and their Metabolite, Butyrate, Improve Experimental Autoimmune Encephalomyelitis
Read this article at
Abstract
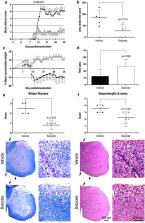
Gut microbiome studies in multiple sclerosis (MS) patients are unravelling some consistent but modest patterns of gut dysbiosis. Among these, a significant decrease of Clostridia cluster IV and XIVa has been reported. In the present study, we investigated the therapeutic effect of a previously selected mixture of human gut-derived 17 Clostridia strains, which belong to Clostridia clusters IV, XIVa, and XVIII, on the clinical outcome of experimental autoimmune encephalomyelitis (EAE). The observed clinical improvement was related to lower demyelination and astrocyte reactivity as well as a tendency to lower microglia reactivity/infiltrating macrophages and axonal damage in the central nervous system (CNS), and to an enhanced immunoregulatory response of regulatory T cells in the periphery. Transcriptome studies also highlighted increased antiinflammatory responses related to interferon beta in the periphery and lower immune responses in the CNS. Since Clostridia-treated mice were found to present higher levels of the immunomodulatory short-chain fatty acid (SCFA) butyrate in the serum, we studied if this clinical effect could be reproduced by butyrate administration alone. Further EAE experiments proved its preventive but slight therapeutic impact on CNS autoimmunity. Thus, this smaller therapeutic effect highlighted that the Clostridia-induced clinical effect was not exclusively related to the SCFA and could not be reproduced by butyrate administration alone. Although it is still unknown if these Clostridia strains will have the same effect on MS patients, gut dysbiosis in MS patients could be partially rebalanced by these commensal bacteria and their immunoregulatory properties could have a beneficial effect on MS clinical course.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found
Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation

- Record: found
- Abstract: found
- Article: found