- Record: found
- Abstract: found
- Article: found
Model‐driven design of a minimal medium for Akkermansia muciniphila confirms mucus adaptation
Read this article at
Summary
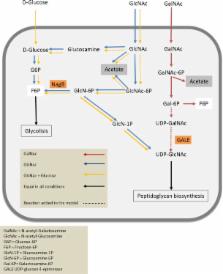
The abundance of the human intestinal symbiont Akkermansia muciniphila has found to be inversely correlated with several diseases, including metabolic syndrome and obesity. A. muciniphila is known to use mucin as sole carbon and nitrogen source. To study the physiology and the potential for therapeutic applications of this bacterium, we designed a defined minimal medium. The composition of the medium was based on the genome‐scale metabolic model of A. muciniphila and the composition of mucin. Our results indicate that A. muciniphila does not code for GlmS, the enzyme that mediates the conversion of fructose‐6‐phosphate (Fru6P) to glucosamine‐6‐phosphate (GlcN6P), which is essential in peptidoglycan formation. The only annotated enzyme that could mediate this conversion is Amuc‐NagB on locus Amuc_1822. We found that Amuc‐NagB was unable to form GlcN6P from Fru6P at physiological conditions, while it efficiently catalyzed the reverse reaction. To overcome this inability, N‐acetylglucosamine needs to be present in the medium for A. muciniphila growth. With these findings, the genome‐scale metabolic model was updated and used to accurately predict growth of A. muciniphila on synthetic media. The finding that A. muciniphila has a necessity for Glc NAc, which is present in mucin further prompts the adaptation to its mucosal niche.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions.

- Record: found
- Abstract: found
- Article: found
COBRApy: COnstraints-Based Reconstruction and Analysis for Python
- Record: found
- Abstract: found
- Article: not found