- Record: found
- Abstract: found
- Article: found
Transcriptome Dynamics of Human Neuronal Differentiation From iPSC
Read this article at
Abstract
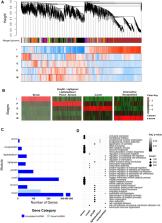
The generation and use of induced pluripotent stem cells (iPSCs) in order to obtain all differentiated adult cell morphologies without requiring embryonic stem cells is one of the most important discoveries in molecular biology. Among the uses of iPSCs is the generation of neuron cells and organoids to study the biological cues underlying neuronal and brain development, in addition to neurological diseases. These iPSC-derived neuronal differentiation models allow us to examine the gene regulatory factors involved in such processes. Among these regulatory factors are long non-coding RNAs (lncRNAs), genes that are transcribed from the genome and have key biological functions in establishing phenotypes, but are frequently not included in studies focusing on protein coding genes. Here, we provide a comprehensive analysis and overview of the coding and non-coding transcriptome during multiple stages of the iPSC-derived neuronal differentiation process using RNA-seq. We identify previously unannotated lncRNAs via genome-guided de novo transcriptome assembly, and the distinct characteristics of the transcriptome during each stage, including differentially expressed and stage specific genes. We further identify key genes of the human neuronal differentiation network, representing novel candidates likely to have critical roles in neurogenesis using coexpression network analysis. Our findings provide a valuable resource for future studies on neuronal differentiation.
Related collections
Most cited references92

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found