- Record: found
- Abstract: found
- Article: found
Transcriptome Comparison of Susceptible and Resistant Wheat in Response to Powdery Mildew Infection

Read this article at
Abstract
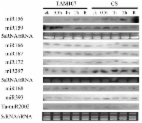
Powdery mildew (Pm) caused by the infection of Blumeria graminis f. sp. tritici ( Bgt) is a worldwide crop disease resulting in significant loss of wheat yield. To profile the genes and pathways responding to the Bgt infection, here, using Affymetrix wheat microarrays, we compared the leaf transcriptomes before and after Bgt inoculation in two wheat genotypes, a Pm-susceptible cultivar Jingdong 8 (S) and its near-isogenic line (R) carrying a single Pm resistant gene Pm30. Our analysis showed that the original gene expression status in the S and R genotypes of wheat was almost identical before Bgt inoculation, since only 60 genes exhibited differential expression by P = 0.01 cutoff. However, 12 h after Bgt inoculation, 3014 and 2800 genes in the S and R genotype, respectively, responded to infection. A wide range of pathways were involved, including cell wall fortification, flavonoid biosynthesis and metabolic processes. Furthermore, for the first time, we show that sense-antisense pair genes might be participants in wheat-powdery mildew interaction. In addition, the results of qRT-PCR analysis on several candidate genes were consistent with the microarray data in their expression patterns. In summary, this study reveals leaf transcriptome changes before and after powdery mildew infection in wheat near-isogenic lines, suggesting that powdery mildew resistance is a highly complex systematic response involving a large amount of gene regulation.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
The lipoxygenase pathway.
- Record: found
- Abstract: found
- Article: not found
The role of abscisic acid in plant-pathogen interactions.
- Record: found
- Abstract: found
- Article: found