- Record: found
- Abstract: found
- Article: found
The regulation of lncRNAs and miRNAs in SARS-CoV-2 infection
Read this article at
Abstract
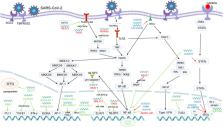
The coronavirus disease 2019 (COVID-19) was a global endemic that continues to cause a large number of severe illnesses and fatalities. There is increasing evidence that non-coding RNAs (ncRNAs) are crucial regulators of viral infection and antiviral immune response and the role of non-coding RNAs in SARS-CoV-2 infection has now become the focus of scholarly inquiry. After SARS-CoV-2 infection, some ncRNAs’ expression levels are regulated to indirectly control the expression of antiviral genes and viral gene replication. However, some other ncRNAs are hijacked by SARS-CoV-2 in order to help the virus evade the immune system by suppressing the expression of type I interferon (IFN-1) and controlling cytokine levels. In this review, we summarize the recent findings of long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) among non-coding RNAs in SARS-CoV-2 infection and antiviral response, discuss the potential mechanisms of actions, and prospects for the detection, treatment, prevention and future directions of SARS-CoV-2 infection research.
Related collections
Most cited references137
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor
- Record: found
- Abstract: found
- Article: not found
Landscape of transcription in human cells
- Record: found
- Abstract: found
- Article: not found