- Record: found
- Abstract: found
- Article: found
Mind the gap: Epigenetic regulation of chromatin accessibility in plants
Read this article at
Abstract
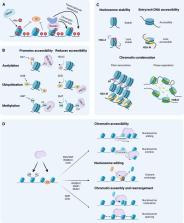
Chromatin plays a crucial role in genome compaction and is fundamental for regulating multiple nuclear processes. Nucleosomes, the basic building blocks of chromatin, are central in regulating these processes, determining chromatin accessibility by limiting access to DNA for various proteins and acting as important signaling hubs. The association of histones with DNA in nucleosomes and the folding of chromatin into higher-order structures are strongly influenced by a variety of epigenetic marks, including DNA methylation, histone variants, and histone post-translational modifications. Additionally, a wide array of chaperones and ATP-dependent remodelers regulate various aspects of nucleosome biology, including assembly, deposition, and positioning. This review provides an overview of recent advances in our mechanistic understanding of how nucleosomes and chromatin organization are regulated by epigenetic marks and remodelers in plants. Furthermore, we present current technologies for profiling chromatin accessibility and organization.
Related collections
Most cited references195
- Record: found
- Abstract: found
- Article: not found
Regulation of chromatin by histone modifications.
- Record: found
- Abstract: found
- Article: not found