- Record: found
- Abstract: found
- Article: found
RiFNet: Automated rib fracture detection in postmortem computed tomography
Read this article at
Abstract
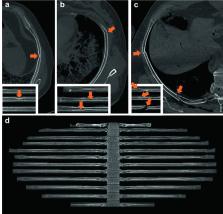
Imaging techniques are widely used for medical diagnostics. In some cases, a lack of medical practitioners who can manually analyze the images can lead to a bottleneck. Consequently, we developed a custom-made convolutional neural network (RiFNet = Rib Fracture Network) that can detect rib fractures in postmortem computed tomography. In a retrospective cohort study, we retrieved PMCT data from 195 postmortem cases with rib fractures from July 2017 to April 2018 from our database. The computed tomography data were prepared using a plugin in the commercial imaging software Syngo.via whereby the rib cage was unfolded on a single-in-plane image reformation. Out of the 195 cases, a total of 585 images were extracted and divided into two groups labeled “with” and “without” fractures. These two groups were subsequently divided into training, validation, and test datasets to assess the performance of RiFNet. In addition, we explored the possibility of applying transfer learning techniques on our dataset by choosing two independent noncommercial off-the-shelf convolutional neural network architectures (ResNet50 V2 and Inception V3) and compared the performances of those two with RiFNet. When using pre-trained convolutional neural networks, we achieved an F 1 score of 0.64 with Inception V3 and an F 1 score of 0.61 with ResNet50 V2. We obtained an average F 1 score of 0.91 ± 0.04 with RiFNet. RiFNet is efficient in detecting rib fractures on postmortem computed tomography. Transfer learning techniques are not necessarily well adapted to make classifications in postmortem computed tomography.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Deep learning.
- Record: found
- Abstract: not found
- Article: not found
Backpropagation Applied to Handwritten Zip Code Recognition
- Record: found
- Abstract: found
- Article: not found