- Record: found
- Abstract: found
- Article: found
Range-wide neutral and adaptive genetic structure of an endemic herb from Amazonian Savannas
Read this article at
Abstract
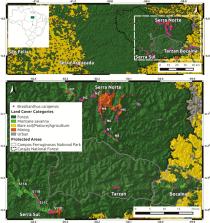
Conserving genetic diversity in rare and narrowly distributed endemic species is essential to maintain their evolutionary potential and minimize extinction risk under future environmental change. In this study we assess neutral and adaptive genetic structure and genetic diversity in Brasilianthus carajensis (Melastomataceae), an endemic herb from Amazonian Savannas. Using RAD sequencing we identified a total of 9365 SNPs in 150 individuals collected across the species’ entire distribution range. Relying on assumption-free genetic clustering methods and environmental association tests we then compared neutral with adaptive genetic structure. We found three neutral and six adaptive genetic clusters, which could be considered management units (MU) and adaptive units (AU), respectively. Pairwise genetic differentiation ( F ST) ranged between 0.024 and 0.048, and even though effective population sizes were below 100, no significant inbreeding was found in any inferred cluster. Nearly 10 % of all analysed sequences contained loci associated with temperature and precipitation, from which only 25 sequences contained annotated proteins, with some of them being very relevant for physiological processes in plants. Our findings provide a detailed insight into genetic diversity, neutral and adaptive genetic structure in a rare endemic herb, which can help guide conservation and management actions to avoid the loss of unique genetic variation.
Abstract
Rare and narrowly distributed plants are particularly sensitive to the loss and fragmentation of their natural habitats, which ultimately result in reduced genetic diversity and a weakened ability to adapt to changing environments. In this study we evaluated the genetic health of a narrowly distributed herb from Amazonian Savannas. Relying on Next-Generation Sequencing technologies we were able to take a detailed glance at a representative portion of the plant’s whole genome, assessing genetic diversity, genetic composition and adaptations to local environmental conditions. Our results provide clear guidance on how to avoid the loss of unique genetic variation in this unique plant.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management.
- Record: found
- Abstract: not found
- Article: not found
Extinction debt of high-mountain plants under twenty-first-century climate change
- Record: found
- Abstract: found
- Article: not found