- Record: found
- Abstract: found
- Article: found
Genetic diversity of Indian garlic core germplasm using agro-biochemical traits and SRAP markers
Read this article at
Abstract
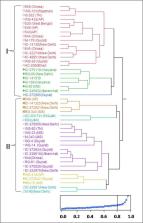
The characterization of garlic germplasm improves its utility, despite the fact that garlic hasn't been used much in the past. Garlic has an untapped genetic pool of immense economic and medicinal value in India. Hence, using heuristic core collection approach, a core set of 46 accessions were selected from 625 Indian garlic accessions based on 13 quantitative and five qualitative traits. The statistical measures (CV per cent, CR per cent, VR per cent) were used to sort the core set using Shannon-Wiener diversity index and the Nei diversity index. In addition, the variation within the core set was tested for 18 agro-morphological and six biochemical characteristics (allicin, phenol content, pyruvic acid, protein, allyl methyl thiosulfinate (AMTHS), and methyl allyl thiosulfinate (MATHS)). Further study of the core set's molecular diversity was performed using sequence related amplified polymorphism (SRAP) markers, which revealed a wide range of diversity among the core set's accessions, with an average polymorphism efficiency (PE) of 80.59 percent, polymorphism information content (PIC) of 0.29, effective multiplex ratio (EMR) of 3.51, and marker index (MI) of 0.99. The findings of this study will be useful in identifying high-yielding, elite garlic germplasm lines with the trait of interest. Since this core set is indicative of total germplasm, these selected breeding lines will be used for genetic improvement of garlic in the future.
Related collections
Most cited references45
- Record: found
- Abstract: not found
- Article: not found
Analysis of Gene Diversity in Subdivided Populations
- Record: found
- Abstract: not found
- Article: not found
Hierarchical Grouping to Optimize an Objective Function
- Record: found
- Abstract: found
- Article: not found