- Record: found
- Abstract: found
- Article: found
Transcriptome profiling of the Caenorhabditis elegans intestine reveals that ELT-2 negatively and positively regulates intestinal gene expression within the context of a gene regulatory network
Read this article at
Abstract
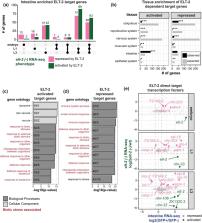
ELT-2 is the major transcription factor (TF) required for Caenorhabditis elegans intestinal development. ELT-2 expression initiates in embryos to promote development and then persists after hatching through the larval and adult stages. Though the sites of ELT-2 binding are characterized and the transcriptional changes that result from ELT-2 depletion are known, an intestine-specific transcriptome profile spanning developmental time has been missing. We generated this dataset by performing Fluorescence Activated Cell Sorting on intestine cells at distinct developmental stages. We analyzed this dataset in conjunction with previously conducted ELT-2 studies to evaluate the role of ELT-2 in directing the intestinal gene regulatory network through development. We found that only 33% of intestine-enriched genes in the embryo were direct targets of ELT-2 but that number increased to 75% by the L3 stage. This suggests additional TFs promote intestinal transcription especially in the embryo. Furthermore, only half of ELT-2's direct target genes were dependent on ELT-2 for their proper expression levels, and an equal proportion of those responded to elt-2 depletion with over-expression as with under-expression. That is, ELT-2 can either activate or repress direct target genes. Additionally, we observed that ELT-2 repressed its own promoter, implicating new models for its autoregulation. Together, our results illustrate that ELT-2 impacts roughly 20–50% of intestine-specific genes, that ELT-2 both positively and negatively controls its direct targets, and that the current model of the intestinal regulatory network is incomplete as the factors responsible for directing the expression of many intestinal genes remain unknown.
Related collections
Most cited references97

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found