- Record: found
- Abstract: found
- Article: found
Mitochondrial HSP70 Chaperone System—The Influence of Post-Translational Modifications and Involvement in Human Diseases
Read this article at
Abstract
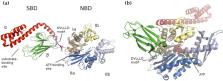
Since their discovery, heat shock proteins (HSPs) have been identified in all domains of life, which demonstrates their importance and conserved functional role in maintaining protein homeostasis. Mitochondria possess several members of the major HSP sub-families that perform essential tasks for keeping the organelle in a fully functional and healthy state. In humans, the mitochondrial HSP70 chaperone system comprises a central molecular chaperone, mtHSP70 or mortalin (HSPA9), which is actively involved in stabilizing and importing nuclear gene products and in refolding mitochondrial precursor proteins, and three co-chaperones (HSP70-escort protein 1—HEP1, tumorous imaginal disc protein 1—TID-1, and Gro-P like protein E—GRPE), which regulate and accelerate its protein folding functions. In this review, we summarize the roles of mitochondrial molecular chaperones with particular focus on the human mtHsp70 and its co-chaperones, whose deregulated expression, mutations, and post-translational modifications are often considered to be the main cause of neurological disorders, genetic diseases, and malignant growth.
Related collections
Most cited references265

- Record: found
- Abstract: found
- Article: found
UniProt: the universal protein knowledgebase in 2021

- Record: found
- Abstract: found
- Article: found
PhosphoSitePlus, 2014: mutations, PTMs and recalibrations
- Record: found
- Abstract: found
- Article: not found