- Record: found
- Abstract: found
- Article: found
Biochemical Convergence of Mitochondrial Hsp70 System Specialized in Iron–Sulfur Cluster Biogenesis
Read this article at
Abstract
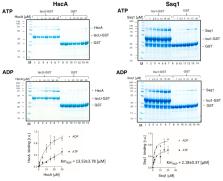
Mitochondria play a central role in the biogenesis of iron–sulfur cluster(s) (FeS), protein cofactors needed for many cellular activities. After assembly on scaffold protein Isu, the cluster is transferred onto a recipient apo-protein. Transfer requires Isu interaction with an Hsp70 chaperone system that includes a dedicated J-domain protein co-chaperone (Hsc20). Hsc20 stimulates Hsp70′s ATPase activity, thus stabilizing the critical Isu–Hsp70 interaction. While most eukaryotes utilize a multifunctional mitochondrial (mt)Hsp70, yeast employ another Hsp70 (Ssq1), a product of mtHsp70 gene duplication. Ssq1 became specialized in FeS biogenesis, recapitulating the process in bacteria, where specialized Hsp70 HscA cooperates exclusively with an ortholog of Hsc20. While it is well established that Ssq1 and HscA converged functionally for FeS transfer, whether these two Hsp70s possess similar biochemical properties was not known. Here, we show that overall HscA and Ssq1 biochemical properties are very similar, despite subtle differences being apparent - the ATPase activity of HscA is stimulated to a somewhat higher levels by Isu and Hsc20, while Ssq1 has a higher affinity for Isu and for Hsc20. HscA/Ssq1 are a unique example of biochemical convergence of distantly related Hsp70s, with practical implications, crossover experimental results can be combined, facilitating understanding of the FeS transfer process.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Causes of molecular convergence and parallelism in protein evolution.
- Record: found
- Abstract: found
- Article: not found
Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity.
- Record: found
- Abstract: found
- Article: not found