- Record: found
- Abstract: found
- Article: found
Fine Analysis of Lymphocyte Subpopulations in SARS-CoV-2 Infected Patients: Differential Profiling of Patients With Severe Outcome

Read this article at
Abstract
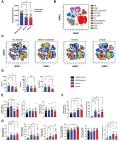
COVID-19 is caused by the human pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and has resulted in widespread morbidity and mortality. CD4 + T cells, CD8 + T cells and neutralizing antibodies all contribute to control SARS-CoV-2 infection. However, heterogeneity is a major factor in disease severity and in immune innate and adaptive responses to SARS-CoV-2. We performed a deep analysis by flow cytometry of lymphocyte populations of 125 hospitalized SARS-CoV-2 infected patients on the day of hospital admission. Five clusters of patients were identified using hierarchical classification on the basis of their immunophenotypic profile, with different mortality outcomes. Some characteristics were observed in all the clusters of patients, such as lymphopenia and an elevated level of effector CD8 +CCR7 - T cells. However, low levels of T cell activation are associated to a better disease outcome; on the other hand, profound CD8 + T-cell lymphopenia, a high level of CD4 + and CD8 + T-cell activation and a high level of CD8 + T-cell senescence are associated with a higher mortality outcome. Furthermore, a cluster of patient was characterized by high B-cell responses with an extremely high level of plasmablasts. Our study points out the prognostic value of lymphocyte parameters such as T-cell activation and senescence and strengthen the interest in treating the patients early in course of the disease with targeted immunomodulatory therapies based on the type of adaptive response of each patient.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: found
Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients

- Record: found
- Abstract: found
- Article: found
Autoantibodies against type I IFNs in patients with life-threatening COVID-19
- Record: found
- Abstract: found
- Article: not found