- Record: found
- Abstract: found
- Article: found
Gene Expression Profiling in Mouse Embryonic Stem Cells Reveals Glycogen Synthase Kinase-3-Dependent Targets of Phosphatidylinositol 3-Kinase and Wnt/β-Catenin Signaling Pathways
Read this article at
Abstract
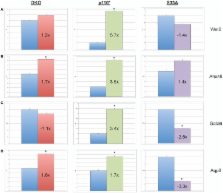
Glycogen synthase kinase-3 (Gsk-3) activity is an important regulator of numerous signal transduction pathways. Gsk-3 activity is the sum of two largely redundant proteins, Gsk-3α and Gsk-3β, and in general, Gsk-3 is a negative regulator of cellular signaling. Genetic deletion of both Gsk-3α and Gsk-3β in mouse embryonic stem cells (ESCs) has previously been shown to lead to the constitutive activation of the Wnt/β-catenin signaling pathway. However, in addition to Wnt signaling, all Gsk-3-regulated pathways, such as insulin signaling, are also affected simultaneously in Gsk-3α − / −; Gsk-3β − / −ESCs. In an effort to better understand how specific signaling pathways contribute to the global pattern of gene expression in Gsk-3α − / −; Gsk-3β − / −ESCs, we compared the gene expression profiles in Gsk-3α − / −; Gsk-3β − / − ESCs to mouse ESCs in which either Wnt/β-catenin signaling or phosphatidylinositol 3-kinase (PI3K)-dependent insulin signaling are constitutively active. Our results show that Wnt signaling has a greater effect on up-regulated genes in the Gsk-3α − / −; Gsk-3β − / −ESCs, whereas PI3K-dependent insulin signaling is more responsible for the down-regulation of genes in the same cells. These data show the importance of Gsk-3 activity on gene expression in mouse ESCs, and that these effects are due to the combined effects of multiple signaling pathways.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: found
In silico prediction of protein-protein interactions in human macrophages
- Record: found
- Abstract: found
- Article: not found
Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B.
- Record: found
- Abstract: found
- Article: not found