- Record: found
- Abstract: found
- Article: found
Promoter Hypomethylation and miR-145-5p Downregulation- Mediated HDAC11 Overexpression Promotes Sorafenib Resistance and Metastasis of Hepatocellular Carcinoma Cells
Read this article at
Abstract
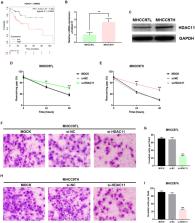
Sorafenib resistance and tumor metastasis account for poor outcome of hepatocellular carcinoma (HCC). Histone deacetylase 11 (HDAC11) has been reported to exert oncogenic effects in several types of human cancer, but its specific functions and detailed mechanisms in HCC are not fully elucidated. Here we identified HDAC11 as a potential oncogene and promising biomarker in HCC by in silico analysis. Histone deacetylase 11 was upregulated in sorafenib-resistant SMMC7721 compared with its parental cell. Knockdown of HDAC11 suppressed proliferation and sorafenib resistance, which may be due to inhibition of drug metabolism cytochrome P450 predicted by gene-set enrichment analysis. Histone deacetylase expression was higher in highly metastatic MHCC97H than lowly metastatic MHCC97L. Downregulation of HDAC11 significantly attenuated the migrated and invaded abilities of HCC cells. Histone deacetylase 11 was directly targeted and suppressed by miR-145-5p. Inhibition of miR-145-5p enhanced sorafenib resistance and metastasis of HCC, and these effects could be attenuated by knockdown of HDAC11. The promoter methylation level of HDAC11 was markedly decreased in HCC tissues compared with normal controls. Administration of 5’-Aza-2’-deoxycytidine, a DNA methyltransferase inhibitor, facilitated HDAC11 expression in HCC cells. Our data indicate a role of miR-145-5p/HDAC11 axis in regulation of sorafenib resistance and metastasis in HCC.
Related collections
Most cited references23

- Record: found
- Abstract: found
- Article: found
circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma

- Record: found
- Abstract: found
- Article: found
Identification of potential miRNA–mRNA regulatory network contributing to pathogenesis of HBV-related HCC
- Record: found
- Abstract: found
- Article: not found