- Record: found
- Abstract: found
- Article: found
NMR Spectroscopy of the Main Protease of SARS‐CoV‐2 and Fragment‐Based Screening Identify Three Protein Hotspots and an Antiviral Fragment

Read this article at
Abstract
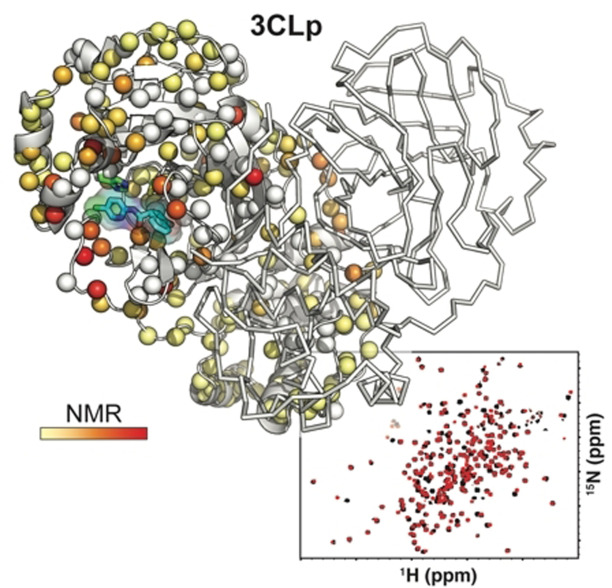
The main protease (3CLp) of the SARS‐CoV‐2, the causative agent for the COVID‐19 pandemic, is one of the main targets for drug development. To be active, 3CLp relies on a complex interplay between dimerization, active site flexibility, and allosteric regulation. The deciphering of these mechanisms is a crucial step to enable the search for inhibitors. In this context, using NMR spectroscopy, we studied the conformation of dimeric 3CLp from the SARS‐CoV‐2 and monitored ligand binding, based on NMR signal assignments. We performed a fragment‐based screening that led to the identification of 38 fragment hits. Their binding sites showed three hotspots on 3CLp, two in the substrate binding pocket and one at the dimer interface. F01 is a non‐covalent inhibitor of the 3CLp and has antiviral activity in SARS‐CoV‐2 infected cells. This study sheds light on the complex structure‐function relationships of 3CLp and constitutes a strong basis to assist in developing potent 3CLp inhibitors.
Abstract
We report the liquid‐sate NMR spectroscopy analysis of the dimeric SARS‐CoV‐2 main protease (3CLp), including its backbone assignments, to study its complex conformational regulation. Using fragment‐based NMR screening, we highlighted three hotspots on the protein, two in the substrate binding pocket and one at the dimer interface, and we identified a non‐covalent reversible inhibitor of 3CLp that has antiviral activity in infected cells.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine

- Record: found
- Abstract: found
- Article: found
A new coronavirus associated with human respiratory disease in China
- Record: found
- Abstract: found
- Article: not found