- Record: found
- Abstract: found
- Article: found
Transcriptomic profiling of taproot growth and sucrose accumulation in sugar beet ( Beta vulgaris L.) at different developmental stages
Read this article at
Abstract
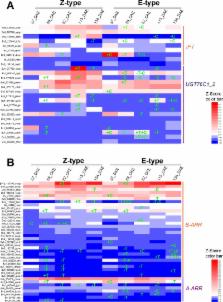
In sugar beet ( Beta vulgaris L.), taproot weight and sucrose content are the important determinants of yield and quality. However, high yield and low sucrose content are two tightly bound agronomic traits. The advances in next-generation sequencing technology and the publication of sugar beet genome have provided a method for the study of molecular mechanism underlying the regulation of these two agronomic traits. In this work, we performed comparative transcriptomic analyses in the high taproot yield cultivar SD13829 and the high sucrose content cultivar BS02 at five developmental stages. More than 50,000,000 pair-end clean reads for each library were generated. When taproot turned into the rapid growth stage at the growth stage of 82 days after emergence (DAE), eighteen enriched gene ontology (GO) terms, including cell wall, cytoskeleton, and enzyme linked receptor protein signaling pathway, occurred in both cultivars. Differentially expressed genes (DEGs) of paired comparison in both cultivars were enriched in the cell wall GO term. For pathway enrichment analyses of DEGs that were respectively generated at 82 DAE compared to 59 DAE (the earlier developmental stage before taproot turning into the rapid growth stage), plant hormone signal transduction pathway was enriched. At 82 DAE, the rapid enlarging stage of taproot, several transcription factor family members were up-regulated in both cultivars. An antagonistic expression of brassinosteroid- and auxin-related genes was also detected. In SD13829, the growth strategy was relatively focused on cell enlargement promoted by brassinosteroid signaling, whereas in BS02, it was relatively focused on secondarily cambial cell division regulated by cytokinin, auxin and brassinosteroid signaling. Taken together, our data demonstrate that the weight and sucrose content of taproot rely on its growth strategy, which is controlled by brassinosteroid, auxin, cytokinin, and gibberellin.
Related collections
Most cited references74

- Record: found
- Abstract: found
- Article: found
RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome
- Record: found
- Abstract: found
- Article: not found
Cluster analysis and display of genome-wide expression patterns.
- Record: found
- Abstract: found
- Article: not found