- Record: found
- Abstract: found
- Article: found
Two New Secreted Proteases Generate a Casein-Derived Antimicrobial Peptide in Bacillus cereus Food Born Isolate Leading to Bacterial Competition in Milk
Read this article at
Abstract
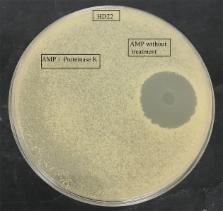
Milk and dairy products harbor a wide variety of bacterial species that compete for both limited resources and space. Under these competitive conditions, bacteria develop specialized mechanisms to protect themselves during niche colonization and nutrient acquisition processes. The bacterial antagonism mechanisms include the production of antimicrobial agents or molecules that facilitate competitor dispersal. In the present work, a bacterial strain designated RC6 was isolated from Ricotta and identified as Bacillus cereus. It generates antimicrobial peptide (AMP) when grown in the presence of casein. The AMP was active against several species of Bacillus and Listeria monocytogenes. MALDI-TOF analysis of the RP-HPLC purified fractions and amino acid sequencing revealed a molecular mass of 751 Da comprised of a 6-residue sequence, YPVEPF. BLAST analysis showed that the AMP corresponds to the fractions 114–119 of bovine β-casein and represents the product of a specific proteolysis. Analysis of the purified proteolytic fractions from the B. cereus RC6 culture supernatant indicated that the presence of at least two different endoproteases is crucial for the generation of the AMP. Indeed, we were able to identify two new candidate endoproteases by means of genome sequencing and functional assignment using a 3D structural model and molecular docking of misannotated hypothetical proteins. In this light, the capacity of B. cereus RC6 to generate antimicrobial peptides from casein, through the production of extracellular enzymes, presents a new model of antagonistic competition leading to niche colonization. Hence, as a dairy product contaminant, this strategy may enable proteolytic B. cereus RC6 niche specialization in milk matrices.
Related collections
Most cited references58
- Record: found
- Abstract: found
- Article: not found
Colicin biology.
- Record: found
- Abstract: found
- Article: not found
Bacteriocins: evolution, ecology, and application.
- Record: found
- Abstract: found
- Article: not found