- Record: found
- Abstract: found
- Article: found
Adenine protonation enables cyclic-di-GMP binding to cyclic-GAMP sensing riboswitches
Read this article at
Abstract
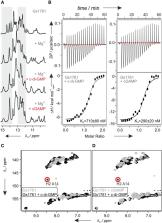
In certain structural or functional contexts, RNA structures can contain protonated nucleotides. However, a direct role for stably protonated nucleotides in ligand binding and ligand recognition has not yet been demonstrated unambiguously. Previous X-ray structures of c-GAMP binding riboswitch aptamer domains in complex with their near-cognate ligand c-di-GMP suggest that an adenine of the riboswitch either forms two hydrogen bonds to a G nucleotide of the ligand in the unusual enol tautomeric form or that the adenine in its N1 protonated form binds the G nucleotide of the ligand in its canonical keto tautomeric state. By using NMR spectroscopy we demonstrate that the c-GAMP riboswitches bind c-di-GMP using a stably protonated adenine in the ligand binding pocket. Thereby, we provide novel insights into the putative biological functions of protonated nucleotides in RNA, which in this case influence the ligand selectivity in a riboswitch.
Related collections
Most cited references60
- Record: found
- Abstract: not found
- Article: not found
Enzymatic assembly of DNA molecules up to several hundred kilobases.
- Record: found
- Abstract: found
- Article: not found
Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence.
- Record: found
- Abstract: found
- Article: not found