- Record: found
- Abstract: found
- Article: not found
Generation of cell-type-specific gene mutations by expressing the sgRNA of the CRISPR system from the RNA polymerase II promoters
research-article
7 June 2015
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Dear Editor,
Recently, the CRISPR/Cas9 system is emerging as a powerful tool for genome editing
(Chang et al., 2013; Li et al., 2013; Niu et al., 2014; Shen et al., 2013; Wan et
al., 2015; Wang et al., 2013) and genetic screening (Konermann et al., 2015), and
holds great promise for biomedical applications in disease modeling and gene therapy
by in vivo genome editing (Maddalo et al., 2014; Xue et al., 2014). However, the applications
of CRISPR/Cas9 system still face some technical hurdles, one of which is to harness
the gene editing in a precisely controlled manner. Of the two-component CRISPR/Cas9
system for genome editing, the Cas9 is a fixed genome-cutting component expressed
from the RNA polymerase II (pol II) promoter that can drive tissue-specific gene expression;
while the single-guide RNA (sgRNA) is a changeable genome-guiding component expressed
from RNA polymerase III (pol III) promoter that usually drives the ubiquitous expression
of “housekeeping” genes in all tissues. Therefore, to express the sgRNA in a tissue-specific
manner can provide a convenient approach to tissue-specific gene mutations. Here,
we reconstructed the sgRNA to enable its expression from the pol II promoters, and
further achieved cell-type specific gene mutations via the modified CRISPR/Cas9 system
by using cell-type specific pol II promoters-driving sgRNA.
To generate pol II promoter-driving sgRNAs, we constructed a microRNA-shRNA-embedded
sgRNA (miRsh-sgRNA) cassette that could express the small RNA from pol II promoter
(Wang et al., 2007) into the 3′-untranslational region (UTR) of the DsRed reporter
gene (Figs. 1A and S1, and Supplementary Materials), as methods by cis-acting ribozymes
(Gao & Zhao, 2014; Nissim et al., 2014) and Cas6/Csy4-based RNA processing (Nissim
et al., 2014) have been reported. Notably, the nonsense shRNAs and a reported efficient
sgRNA targeting the mouse p53 gene (sgp53) (Xue et al., 2014) was adopted in the construct
for a proof-of-concept experiment. The mature sgRNA derived from the miRsh-sgRNA cassette
will have additive 7 nucleotides in the 5′-end (Fig. 1A), so we chose a reported optimized
sgRNA backbone, the sgRNA(F+E), to improve the mutagenesis efficiency (Chen et al.,
2013), given that the sgRNAs with mispairing and addition in the 5′-end are still
functional (Cong et al., 2013).
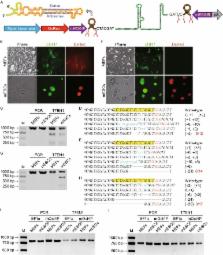
Figure 1
Pol II promoter-driving miRsh-sgp53 functions in MEFs and mESCs. (A) Structure of
pol II promoter-driving miRsh-sgRNA cassette. The cut sites of Drasha in this primary-microRNA
are marked, and the resulted sgRNA would have 7 additive nucleotides (CTACGAT) at
the 5′-end. (B) MEFs and mESCs transfected with Cas9-EGFP and constitutive EF1a promoter-driving
DsRed-miRsh-sgp53 vectors are both GFP and DsRed positive at 48 h after transfection.
(C) T7EN1 cleavage assay showed that constitutive EF1a promoter-driving miRsh-sgp53
can guide Cas9 for producing DSBs in both MEFs and mESCs. The PCR band is 972 bp and
the theoretical cut bands are 286 bp and 686 bp. (D and E) Representative Sanger sequencing
results of the PCR amplicons from MEFs (D) or mESCs (E) transfected with EF1a-driving
miRsh-sgp53 cassette. AGG (red) is the protospacer-adjacent motif (PAM) sequence.
Mutations were described in brackets. (F) MEFs transfected with Cas9-EGFP and stem
cell-specific mOct4P promoter-driving DsRed-miRsh-sgp53 vectors are only GFP positive
while mESCs are both GFP and DsRed positive at 48 h after transfection. (G) T7EN1
cleavage assay showed that stem-cell-specific mOct4P promoter-driving miRsh-sgp53
can guide Cas9 for producing DSBs only in mESCs but not in MEFs. The PCR band is 972
bp and the theoretical cut bands are 286 bp and 686 bp. (H) Representative Sanger
sequencing results of the PCR amplicons from mESCs transfected with mOct4P-driving
miRsh-sgp53 cassette. AGG (red) is the protospacer-adjacent motif (PAM) sequence.
Mutations were described in brackets. (I) Off targeting analysis of p53-off-27 site
in MEFs and mESCs transfected with EF1a-driving miRsh-sgp53 and Cas9 or mOct4P-driving
miRsh-sgp53 and Cas9 by T7EN1 assay. The PCR band is 800 bp and the theoretical cut
bands are 315 bp and 485 bp. (J) Off targeting analysis of p53-off-28 site. The PCR
band is 753 bp and the theoretical cut bands are 281 bp and 472 bp
To test whether functional sgRNA can be efficiently derived from the pol II promoter-driving
miRsh-sgRNA cassette, we transfected mouse embryonic fibroblasts (MEFs) and mouse
embryonic stem cells (mESCs) with constitutive EF1a promoter-driving miRsh-sgp53 expression
vector and Cas9-P2A-EGFP expression vector. GFP and DsRed double positive cells were
sorted by fluorescence-activated cell sorting (FACS) two days after transfection for
further analysis (Fig. 1B). The T7EN1 cleavage assay of these cells showed that constitutive
EF1a promoter-driving miRsh-sgp53 can guide Cas9 for producing double strand breaks
(DSBs) in p53 gene in both MEFs and mESCs (Fig. 1C). To confirm the T7EN1 cleavage
results, Sanger sequencing was performed and the result showed that the efficiency
of miRsh-sgp53 in MEFs (Fig. 1D) and mESCs (Fig. 1E) was about 50% (6/12) and 57.1%
(8/14). These results indicated that the functional sgRNA could be expressed from
the pol II promoter-driving miRsh-sgRNA construct for successful gene mutations.
Further, to examine whether the miRsh-sgRNA cassette can produce gene mutation in
a cell type-specific manner, we constructed an expression vector using the embryonic
stem cell-specific mouse Oct4 gene promoter (mOct4P) to express the miRsh-sgp53 cassette.
Two days after transfection of MEFs and mESCs with this vector and the EF1a promoter-driving
Cas9-P2A-EGFP vector, GFP and DsRed double-positive mESCs were observed, but only
GFP positive MEFs were observed, indicating the cell-type-specific expression of the
sgRNA (Fig. 1F). We sorted GFP and DsRed double positive mESCs and GFP positive MEFs
by FACS for further analysis. T7EN1 cleavage assay and Sanger sequencing were performed,
and p53 gene mutation was only detected in the mESCs but not in the MEFs (Fig. 1G).
The result of Sanger sequencing showed that the p53 mutation efficiency in mESCs with
ESC-specific mOct4P-driving miRsh-sgp53 (52.9%, 9/17) was similar to that with constitutive
EF1a promoter-driving sgp53 (Fig. 1H). These results indicated that the cell type-specific
gene editing can be achieved by cell type-specific promoter-driving expression of
miRsh-sgRNA.
Previous studies suggested that CRISPR/Cas9 system would probably induce off-target
mutations because the binding to genome could tolerate sequence mismatches distal
from the PAM at the 5′ end of sgRNAs. It has been demonstrated in bacteria and cultured
human cells that the DNA cleavage specificity of CRISPR/Cas9 system is determined
by the PAM sequence NGG and the 8–12 base ‘‘seed sequence’’ at the 3′ end of the sgRNA
(Cong et al., 2013; Jinek et al., 2013; Wang et al., 2013). We searched for potential
off target sites based on this rule, and found 41 potential off targets of the sgp53
(named p53-off-1 to p53-off-41) existing in mouse genome (Table S2). The T7EN1 assay
and Sanger sequencing revealed that no potential off target site we tested (Figs. 1I,
1J and S2) was mutated by miRsh-sgp53 in all the examined cells.
In conclusion, we designed a new construct for efficient and visible expression of
sgRNAs from the pol II promoters, which therefore can produce cell-type specific mutations.
This reconstructed pol II promoter-driving miRsh-sgRNA backbone will make the CRISPR/Cas9
system-mediated genome editing be more controllable and safer for future applications
such as in in vivo gene therapy.
Electronic supplementary material
Supplementary material 1 (PDF 958 kb)
Related collections
Most cited references5
- Record: found
- Abstract: found
- Article: not found
CRISPR-mediated direct mutation of cancer genes in the mouse liver
Wen-Bin Xue, Sidi Chen, Hao Yin … (2014)
- Record: found
- Abstract: not found
- Article: not found
Generation of gene-modified mice via Cas9/RNA-mediated gene targeting.
- Record: found
- Abstract: found
- Article: not found
Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing.
Yangbin Gao, Yunde Zhao (2014)
