- Record: found
- Abstract: found
- Article: found
Long noncoding RNA ENST00000436340 promotes podocyte injury in diabetic kidney disease by facilitating the association of PTBP1 with RAB3B
Read this article at
Abstract
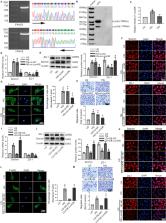
Dysfunction of podocytes has been regarded as an important early pathologic characteristic of diabetic kidney disease (DKD), but the regulatory role of long noncoding RNAs (lncRNAs) in this process remains largely unknown. Here, we performed RNA sequencing in kidney tissues isolated from DKD patients and nondiabetic renal cancer patients undergoing surgical resection and discovered that the novel lncRNA ENST00000436340 was upregulated in DKD patients and high glucose-induced podocytes, and we showed a significant correlation between ENST00000436340 and kidney injury. Gain- and loss-of-function experiments showed that silencing ENST00000436340 alleviated high glucose-induced podocyte injury and cytoskeleton rearrangement. Mechanistically, we showed that fat mass and obesity- associate gene (FTO)-mediated m6A induced the upregulation of ENST00000436340. ENST00000436340 interacted with polypyrimidine tract binding protein 1 (PTBP1) and augmented PTBP1 binding to RAB3B mRNA, promoted RAB3B mRNA degradation, and thereby caused cytoskeleton rearrangement and inhibition of GLUT4 translocation to the plasma membrane, leading to podocyte injury and DKD progression. Together, our results suggested that upregulation of ENST00000436340 could promote podocyte injury through PTBP1-dependent RAB3B regulation, thus suggesting a novel form of lncRNA-mediated epigenetic regulation of podocytes that contributes to the pathogenesis of DKD.
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
Long non-coding RNAs: new players in cell differentiation and development.
- Record: found
- Abstract: found
- Article: not found
m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer.
- Record: found
- Abstract: found
- Article: found