- Record: found
- Abstract: found
- Article: found
CRISPR/Cas9-mediated excision of ALS/FTD-causing hexanucleotide repeat expansion in C9ORF72 rescues major disease mechanisms in vivo and in vitro
Read this article at
Abstract
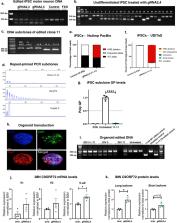
A GGGGCC 24+ hexanucleotide repeat expansion (HRE) in the C9ORF72 gene is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), fatal neurodegenerative diseases with no cure or approved treatments that substantially slow disease progression or extend survival. Mechanistic underpinnings of neuronal death include C9ORF72 haploinsufficiency, sequestration of RNA-binding proteins in the nucleus, and production of dipeptide repeat proteins. Here, we used an adeno-associated viral vector system to deliver CRISPR/Cas9 gene-editing machineries to effectuate the removal of the HRE from the C9ORF72 genomic locus. We demonstrate successful excision of the HRE in primary cortical neurons and brains of three mouse models containing the expansion (500–600 repeats) as well as in patient-derived iPSC motor neurons and brain organoids (450 repeats). This resulted in a reduction of RNA foci, poly-dipeptides and haploinsufficiency, major hallmarks of C9-ALS/FTD, making this a promising therapeutic approach to these diseases.
Abstract
A hexanucleotide repeat expansion in C9ORF72 is the most common genetic cause of ALS and FTD. Here, the authors demonstrate CRISPR/Cas9 excision of the expansion results in a rescue of disease mechanisms in vivo and in vitro.
Related collections
Most cited references80
- Record: found
- Abstract: found
- Article: not found
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.
- Record: found
- Abstract: found
- Article: not found
Multiplex genome engineering using CRISPR/Cas systems.
- Record: found
- Abstract: found
- Article: not found