- Record: found
- Abstract: found
- Article: found
The Colletotrichum gloeosporioides species complex
Read this article at
Abstract
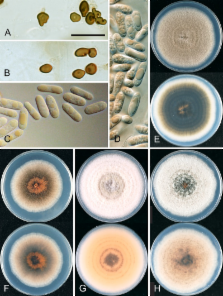
The limit of the Colletotrichum gloeosporioides species complex is defined genetically, based on a strongly supported clade within the Colletotrichum ITS gene tree. All taxa accepted within this clade are morphologically more or less typical of the broadly defined C. gloeosporioides, as it has been applied in the literature for the past 50 years. We accept 22 species plus one subspecies within the C. gloeosporioides complex. These include C. asianum, C. cordylinicola, C. fructicola, C. gloeosporioides, C. horii, C. kahawae subsp. kahawae, C. musae, C. nupharicola, C. psidii, C. siamense, C. theobromicola, C. tropicale, and C. xanthorrhoeae, along with the taxa described here as new, C. aenigma, C. aeschynomenes, C. alatae, C. alienum, C. aotearoa, C. clidemiae, C. kahawae subsp. ciggaro, C. salsolae, and C. ti, plus the nom. nov. C. queenslandicum (for C. gloeosporioides var. minus). All of the taxa are defined genetically on the basis of multi-gene phylogenies. Brief morphological descriptions are provided for species where no modern description is available. Many of the species are unable to be reliably distinguished using ITS, the official barcoding gene for fungi. Particularly problematic are a set of species genetically close to C. musae and another set of species genetically close to C. kahawae, referred to here as the Musae clade and the Kahawae clade, respectively. Each clade contains several species that are phylogenetically well supported in multi-gene analyses, but within the clades branch lengths are short because of the small number of phylogenetically informative characters, and in a few cases individual gene trees are incongruent. Some single genes or combinations of genes, such as glyceraldehyde-3-phosphate dehydrogenase and glutamine synthetase, can be used to reliably distinguish most taxa and will need to be developed as secondary barcodes for species level identification, which is important because many of these fungi are of biosecurity significance. In addition to the accepted species, notes are provided for names where a possible close relationship with C. gloeosporioides sensu lato has been suggested in the recent literature, along with all subspecific taxa and formae speciales within C. gloeosporioides and its putative teleomorph Glomerella cingulata.
Taxonomic novelties:
Name replacement - C. queenslandicum B. Weir & P.R. Johnst. New species - C. aenigma B. Weir & P.R. Johnst., C. aeschynomenes B. Weir & P.R. Johnst., C. alatae B. Weir & P.R. Johnst., C . alienum B. Weir & P.R. Johnst, C. aotearoa B. Weir & P.R. Johnst., C. clidemiae B. Weir & P.R. Johnst., C. salsolae B. Weir & P.R. Johnst., C. ti B. Weir & P.R. Johnst. New subspecies - C. kahawae subsp. ciggaro B. Weir & P.R. Johnst. Typification: Epitypification - C. queenslandicum B. Weir & P.R. Johnst.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Phylogenetic species recognition and species concepts in fungi.
- Record: found
- Abstract: found
- Article: found
A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis
- Record: found
- Abstract: found
- Article: not found