- Record: found
- Abstract: found
- Article: found
Influence on the fermentation quality, microbial diversity, and metabolomics in the ensiling of sunflower stalks and alfalfa
Read this article at
Abstract
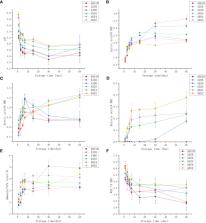
With the rapid development of the livestock industry, finding new sources of feed has become a critical issue that needs to be addressed urgently. China is one of the top five sunflower producers in the world and generates a massive amount of sunflower stalks annually, yet this resource has not been effectively utilized. Therefore, in order to tap into the potential of sunflower stalks for animal feed, it is essential to explore and develop efficient methods for their utilization.In this study, various proportions of alfalfa and sunflower straw were co-ensiled with the following mixing ratios: 0:10, 2:8, 4:6, 5:5, 6:4, and 8:2, denoted as A0S10, A2S8, A4S6, A5S5, A6S4, and A8S2, respectively. The nutrient composition, fermentation quality, microbial quantity, microbial diversity, and broad-spectrum metabolomics on the 60th day were assessed. The results showed that the treatment groups with more sunflower straw added (A2S8, A4S6) could start fermentation earlier. On the first day of fermentation, Weissella spp.dominated overwhelmingly in these two groups. At the same time, in the early stage of fermentation, the pH in these two groups dropped rapidly, which could effectively reduce the loss of nutrients in the early stage of fermentation.In the later fermentation period, a declining trend in acetic acid levels was observed in A0S10, A2S8, and A4S6, while no butyric acid production was detected in A0S10 and A2S8 throughout the process. In A4S6, butyric acid production was observed only after 30 days of fermentation. From the perspective of metabolites, compared with sunflower ensiling alone, many bioactive substances such as flavonoids, alkaloids, and terpenes are upregulated in mixed ensiling.
Related collections
Most cited references61

- Record: found
- Abstract: found
- Article: found
fastp: an ultra-fast all-in-one FASTQ preprocessor
- Record: found
- Abstract: found
- Article: not found
FLASH: fast length adjustment of short reads to improve genome assemblies.
- Record: found
- Abstract: found
- Article: not found