- Record: found
- Abstract: found
- Article: found
Transcriptional Profiling in Experimental Visceral Leishmaniasis Reveals a Broad Splenic Inflammatory Environment that Conditions Macrophages toward a Disease-Promoting Phenotype
Read this article at
Abstract
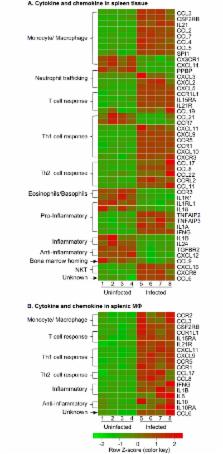
Visceral Leishmaniasis (VL), caused by the intracellular protozoan Leishmania donovani, is characterized by relentlessly increasing visceral parasite replication, cachexia, massive splenomegaly, pancytopenia and ultimately death. Progressive disease is considered to be due to impaired effector T cell function and/or failure of macrophages to be activated to kill the intracellular parasite. In previous studies, we used the Syrian hamster ( Mesocricetus auratus) as a model because it mimics the progressive nature of active human VL. We demonstrated previously that mixed expression of macrophage-activating (IFN-γ) and regulatory (IL-4, IL-10, IL-21) cytokines, parasite-induced expression of macrophage arginase 1 (Arg1), and decreased production of nitric oxide are key immunopathologic factors. Here we examined global changes in gene expression to define the splenic environment and phenotype of splenic macrophages during progressive VL. We used RNA sequencing coupled with de novo transcriptome assembly, because the Syrian hamster does not have a fully sequenced and annotated reference genome. Differentially expressed transcripts identified a highly inflammatory spleen environment with abundant expression of type I and type II interferon response genes. However, high IFN-γ expression was ineffective in directing exclusive M1 macrophage polarization, suppressing M2-associated gene expression, and restraining parasite replication and disease. While many IFN-inducible transcripts were upregulated in the infected spleen, fewer were induced in splenic macrophages in VL. Paradoxically, IFN-γ enhanced parasite growth and induced the counter-regulatory molecules Arg1, Ido1 and Irg1 in splenic macrophages. This was mediated, at least in part, through IFN-γ-induced activation of STAT3 and expression of IL-10, which suggests that splenic macrophages in VL are conditioned to respond to macrophage activation signals with a counter-regulatory response that is ineffective and even disease-promoting. Accordingly, inhibition of STAT3 activation led to a reduced parasite load in infected macrophages. Thus, the STAT3 pathway offers a rational target for adjunctive host-directed therapy to interrupt the pathogenesis of VL.
Author Summary
Visceral leishmaniasis (VL) is a neglected parasitic disease that is caused by the intracellular protozoan Leishmania donovani. Patients with this disease suffer from muscle wasting, enlargement of the spleen, reduced blood counts and ultimately will die without treatment. Progressive disease is considered to be due to impaired cellular immunity, with T cell or macrophage dysfunction, or both. We studied the Syrian hamster as an infection model because it mimics the progressive nature of human disease. We examined global changes in gene expression in the spleen and splenic macrophages during experimental VL and identified a highly inflammatory spleen environment with abundant expression of interferon and interferon-response genes that would be expected to control the infection. However, the high level of IFN-γ expression was ineffective in mediating a protective macrophage response, restraining parasite replication and halting progression of disease. We found that IFN-γ itself stimulated parasite growth in splenic macrophages and induced expression of counter-regulatory molecules, which may paradoxically make the host more susceptible. These data give insights into the nature of the immune response that promotes the infection, and identifies potential targets for therapeutic intervention.
Related collections
Most cited references95
- Record: found
- Abstract: found
- Article: not found
Small-sample estimation of negative binomial dispersion, with applications to SAGE data.
- Record: found
- Abstract: found
- Article: not found
IRF7: activation, regulation, modification and function.
- Record: found
- Abstract: found
- Article: not found