- Record: found
- Abstract: found
- Article: found
Single-Cell Network Analysis Identifies DDIT3 as a Nodal Lineage Regulator in Hematopoiesis

Read this article at
Summary
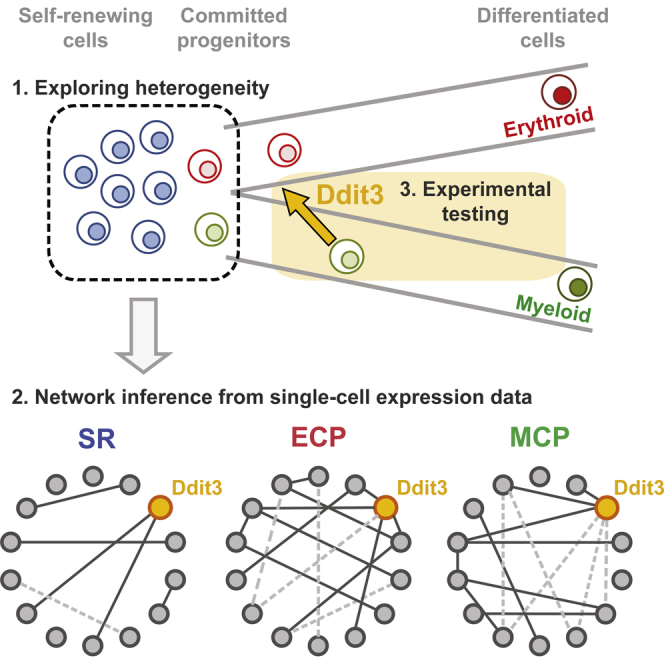
We explore cell heterogeneity during spontaneous and transcription-factor-driven commitment for network inference in hematopoiesis. Since individual genes display discrete OFF states or a distribution of ON levels, we compute and combine pairwise gene associations from binary and continuous components of gene expression in single cells. Ddit3 emerges as a regulatory node with positive linkage to erythroid regulators and negative association with myeloid determinants. Ddit3 loss impairs erythroid colony output from multipotent cells, while forcing Ddit3 in granulo-monocytic progenitors (GMPs) enhances self-renewal and impedes differentiation. Network analysis of Ddit3-transduced GMPs reveals uncoupling of myeloid networks and strengthening of erythroid linkages. RNA sequencing suggests that Ddit3 acts through development or stabilization of a precursor upstream of GMPs with inherent Meg-E potential. The enrichment of Gata2 target genes in Ddit3-dependent transcriptional responses suggests that Ddit3 functions in an erythroid transcriptional network nucleated by Gata2.
Graphical Abstract
Highlights
-
•
We present a method for inferring gene regulatory networks (GRNs) from single cells
-
•
Lineage cross-antagonism is a key property of GRNs of early lineage commitment
-
•
Ddit3 is a regulatory node in erythroid lineage programming
-
•
A Ddit3-Gata2 regulatory axis antagonizes myeloid and enables erythroid programs
Abstract
Pina et al. develop a gene regulatory network inference method using single-cell gene expression data and identify Ddit3 as a regulatory node in erythroid lineage programming. The authors explore this inference and show that Ddit3 can antagonize myeloid programming and enable erythroid signatures and forms a regulatory axis with Gata2.
Related collections
Most cited references15
- Record: found
- Abstract: found
- Article: not found
CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum.
- Record: found
- Abstract: found
- Article: not found
Forcing cells to change lineages.
- Record: found
- Abstract: found
- Article: not found