- Record: found
- Abstract: found
- Article: found
Viral infections at the animal–human interface—Learning lessons from the SARS‐CoV‐2 pandemic
Read this article at
Abstract
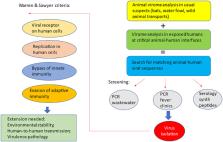
This Lilliput explores the current epidemiological and virological arguments for a zoonotic origin of the COVID‐19 pandemic. While the role of bats, pangolins and racoon dogs as viral reservoirs has not yet been proven, a spill‐over of a coronavirus infection from animals into humans at the Huanan food market in Wuhan has a much greater plausibility than alternative hypotheses such as a laboratory virus escape, deliberate genetic engineering or introduction by cold chain food products. This Lilliput highlights the dynamic nature of the animal‐human interface for viral cross‐infections from humans into feral white tail deer or farmed minks (reverse zoonosis). Surveillance of viral infections at the animal‐human interface is an urgent task since live animal markets are not the only risks for future viral spill‐overs. Climate change will induce animal migration which leads to viral exchanges between animal species that have not met in the past. Environmental change and deforestation will also increase contact between animals and humans. Developing an early warning system for emerging viral infections becomes thus a societal necessity not only for human but also for animal and environmental health (One Health concept). Microbiologists have developed tools ranging from virome analysis in key suspects such as viral reservoirs (bats, wild game animals, bushmeat) and in humans exposed to wild animals, to wastewater analysis to detect known and unknown viruses circulating in the human population and sentinel studies in animal‐exposed patients with fever. Criteria need to be developed to assess the virulence and transmissibility of zoonotic viruses. An early virus warning system is costly and will need political lobbying. The accelerating number of viral infections with pandemic potential over the last decades should provide the public pressure to extend pandemic preparedness for the inclusion of early viral alert systems.
Related collections
Most cited references72

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
Global trends in emerging infectious diseases
- Record: found
- Abstract: found
- Article: not found