- Record: found
- Abstract: found
- Article: found
Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer
Read this article at
SUMMARY
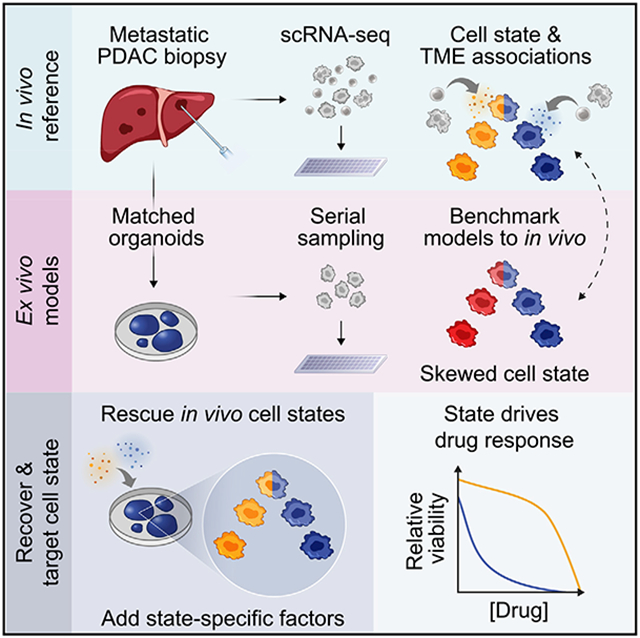
Prognostically relevant RNA expression states exist in pancreatic ductal adenocarcinoma (PDAC), but our understanding of their drivers, stability, and relationship to therapeutic response is limited. To examine these attributes systematically, we profiled metastatic biopsies and matched organoid models at single-cell resolution. In vivo, we identify a new intermediate PDAC transcriptional cell state and uncover distinct site- and state-specific tumor microenvironments (TMEs). Benchmarking models against this reference map, we reveal strong culture-specific biases in cancer cell transcriptional state representation driven by altered TME signals. We restore expression state heterogeneity by adding back in vivo-relevant factors and show plasticity in culture models. Further, we prove that non-genetic modulation of cell state can strongly influence drug responses, uncovering state-specific vulnerabilities. This work provides a broadly applicable framework for aligning cell states across in vivo and ex vivo settings, identifying drivers of transcriptional plasticity and manipulating cell state to target associated vulnerabilities.
Graphical Abstract
In brief
Systematic profiling of metastatic pancreatic cancer biopsies and matched organoid models provides a view of cellular states, their regulation by the tumor microenvironment, and the ability to modulate these states to impact drug responses.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
The Cancer Genome Atlas Pan-Cancer analysis project.
- Record: found
- Abstract: found
- Article: not found
Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets.
- Record: found
- Abstract: found
- Article: not found
Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.