- Record: found
- Abstract: found
- Article: found
Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
Read this article at
Abstract
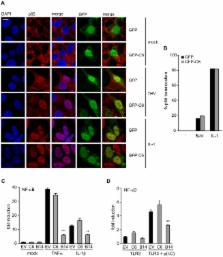
Recognition of viruses by pattern recognition receptors (PRRs) causes interferon-β (IFN-β) induction, a key event in the anti-viral innate immune response, and also a target of viral immune evasion. Here the vaccinia virus (VACV) protein C6 is identified as an inhibitor of PRR-induced IFN-β expression by a functional screen of select VACV open reading frames expressed individually in mammalian cells. C6 is a member of a family of Bcl-2-like poxvirus proteins, many of which have been shown to inhibit innate immune signalling pathways. PRRs activate both NF-κB and IFN regulatory factors (IRFs) to activate the IFN-β promoter induction. Data presented here show that C6 inhibits IRF3 activation and translocation into the nucleus, but does not inhibit NF-κB activation. C6 inhibits IRF3 and IRF7 activation downstream of the kinases TANK binding kinase 1 (TBK1) and IκB kinase-ε (IKKε), which phosphorylate and activate these IRFs. However, C6 does not inhibit TBK1- and IKKε-independent IRF7 activation or the induction of promoters by constitutively active forms of IRF3 or IRF7, indicating that C6 acts at the level of the TBK1/IKKε complex. Consistent with this notion, C6 immunoprecipitated with the TBK1 complex scaffold proteins TANK, SINTBAD and NAP1. C6 is expressed early during infection and is present in both nucleus and cytoplasm. Mutant viruses in which the C6L gene is deleted, or mutated so that the C6 protein is not expressed, replicated normally in cell culture but were attenuated in two in vivo models of infection compared to wild type and revertant controls. Thus C6 contributes to VACV virulence and might do so via the inhibition of PRR-induced activation of IRF3 and IRF7.
Author Summary
A key event in the innate immune response to virus infection is the detection of pathogen-associated molecular patterns (PAMPs) such as viral DNA and RNA by cellular pattern recognition receptors (PRRs). This leads to expression of interferon-β (IFN-β) by an infected cell. Many viruses have evolved mechanisms to evade the induction of IFN-β. Here a screen of poorly characterized vaccinia virus (VACV) proteins identified protein C6 as an inhibitor of IFN-β induction by PRRs. Data presented show that C6 prevents the activation of the transcription factors IRF3 and IRF7 by the kinases TBK1 and IKKε, which are key components at the point of convergence of several PRR signalling pathways. C6 interacts with the scaffold proteins NAP1, TANK and SINTBAD, which are components of the protein complexes containing TBK1 and IKKε, and this interaction might modulate the activity of these kinases. C6 is expressed early during infection and contributes to virulence because viruses that do not express C6 are attenuated in two in vivo models compared to wild type and revertant control viruses.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates.
- Record: found
- Abstract: found
- Article: not found
Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures.
- Record: found
- Abstract: found
- Article: not found