- Record: found
- Abstract: found
- Article: not found
Understanding COVID-19-associated coagulopathy
Read this article at
Abstract
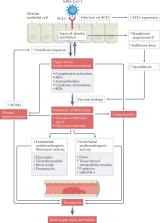
COVID-19-associated coagulopathy (CAC) is a life-threatening complication of SARS-CoV-2 infection. However, the underlying cellular and molecular mechanisms driving this condition are unclear. Evidence supports the concept that CAC involves complex interactions between the innate immune response, the coagulation and fibrinolytic pathways, and the vascular endothelium, resulting in a procoagulant condition. Understanding of the pathogenesis of this condition at the genomic, molecular and cellular levels is needed in order to mitigate thrombosis formation in at-risk patients. In this Perspective, we categorize our current understanding of CAC into three main pathological mechanisms: first, vascular endothelial cell dysfunction; second, a hyper-inflammatory immune response; and last, hypercoagulability. Furthermore, we pose key questions and identify research gaps that need to be addressed to better understand CAC, facilitate improved diagnostics and aid in therapeutic development. Finally, we consider the suitability of different animal models to study CAC.
Abstract
Here, the authors consider our emerging understanding of COVID-19-associated coagulopathy. They focus on the complex interactions between innate immune, coagulation and fibrinolytic pathways that can lead to potentially life-threatening thrombosis following SARS-CoV-2 infection.
Related collections
Most cited references163
- Record: found
- Abstract: found
- Article: not found
Clinical Characteristics of Coronavirus Disease 2019 in China
- Record: found
- Abstract: found
- Article: not found
Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study
- Record: found
- Abstract: found
- Article: not found
