- Record: found
- Abstract: found
- Article: found
Chitin Synthases from Saprolegnia Are Involved in Tip Growth and Represent a Potential Target for Anti-Oomycete Drugs
Read this article at
Abstract
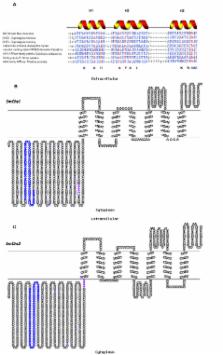
Oomycetes represent some of the most devastating plant and animal pathogens. Typical examples are Phytophthora infestans, which causes potato and tomato late blight, and Saprolegnia parasitica, responsible for fish diseases. Despite the economical and environmental importance of oomycete diseases, their control is difficult, particularly in the aquaculture industry. Carbohydrate synthases are vital for hyphal growth and represent interesting targets for tackling the pathogens. The existence of 2 different chitin synthase genes ( SmChs1 and SmChs2) in Saprolegnia monoica was demonstrated using bioinformatics and molecular biology approaches. The function of SmCHS2 was unequivocally demonstrated by showing its catalytic activity in vitro after expression in Pichia pastoris. The recombinant SmCHS1 protein did not exhibit any activity in vitro, suggesting that it requires other partners or effectors to be active, or that it is involved in a different process than chitin biosynthesis. Both proteins contained N-terminal Microtubule Interacting and Trafficking domains, which have never been reported in any other known carbohydrate synthases. These domains are involved in protein recycling by endocytosis. Enzyme kinetics revealed that Saprolegnia chitin synthases are competitively inhibited by nikkomycin Z and quantitative PCR showed that their expression is higher in presence of the inhibitor. The use of nikkomycin Z combined with microscopy showed that chitin synthases are active essentially at the hyphal tips, which burst in the presence of the inhibitor, leading to cell death. S. parasitica was more sensitive to nikkomycin Z than S. monoica. In conclusion, chitin synthases with species-specific characteristics are involved in tip growth in Saprolegnia species and chitin is vital for the micro-organisms despite its very low abundance in the cell walls. Chitin is most likely synthesized transiently at the apex of the cells before cellulose, the major cell wall component in oomycetes. Our results provide important fundamental information on cell wall biogenesis in economically important species, and demonstrate the potential of targeting oomycete chitin synthases for disease control.
Author Summary
Oomycete pathogens can infect many organisms relevant to the agriculture and aquaculture industries, such as potato and tomato, or fishes like salmon. Saprolegnia parasitica represents the most important oomycete fish pathogen that challenges the productivity of fish farms due to the lack of efficient methods for containing its development and pathogenicity. Enzymes involved in cell wall formation represent potential targets of anti-oomycete drugs. The isolation and full characterization of two genes involved in the biosynthesis of chitin, a quantitatively minor cell wall carbohydrate in Saprolegnia, was performed. Despite its low abundance, chitin was shown to play a key role in hyphal tip growth, which is a vital process for the micro-organism. The enzymes responsible for chitin biosynthesis were located at the apex of the hyphae and specifically inhibited by nikkomycin Z. The inhibitor provoked cell death by bursting of the hyphal tips. S. parasitica was more sensitive to the inhibitor than the model species Saprolegnia monoica used for these investigations. The data demonstrate the potential of targeting chitin synthases to control the diseases caused by S. parasitica and pave the way for the establishment of sustainable methods to tackle the adverse effects of the pathogen.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
ProtTest: selection of best-fit models of protein evolution.
- Record: found
- Abstract: found
- Article: not found
Rapid isolation of high molecular weight plant DNA.
- Record: found
- Abstract: found
- Article: not found