- Record: found
- Abstract: found
- Article: found
Application of text mining to develop AOP-based mucus hypersecretion genesets and confirmation with in vitro and clinical samples
Read this article at
Abstract
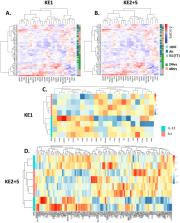
Mucus hypersecretion contributes to lung function impairment observed in COPD (chronic obstructive pulmonary disease), a tobacco smoking-related disease. A detailed mucus hypersecretion adverse outcome pathway (AOP) has been constructed from literature reviews, experimental and clinical data, mapping key events (KEs) across biological organisational hierarchy leading to an adverse outcome. AOPs can guide the development of biomarkers that are potentially predictive of diseases and support the assessment frameworks of nicotine products including electronic cigarettes. Here, we describe a method employing manual literature curation supported by a focused automated text mining approach to identify genes involved in 5 KEs contributing to decreased lung function observed in tobacco-related COPD. KE genesets were subsequently confirmed by unsupervised clustering against 3 different transcriptomic datasets including (1) in vitro acute cigarette smoke and e-cigarette aerosol exposure, (2) in vitro repeated incubation with IL-13, and (3) lung biopsies from COPD and healthy patients. The 5 KE genesets were demonstrated to be predictive of cigarette smoke exposure and mucus hypersecretion in vitro, and less conclusively predict the COPD status of lung biopsies. In conclusion, using a focused automated text mining and curation approach with experimental and clinical data supports the development of risk assessment strategies utilising AOPs.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
KEGG: kyoto encyclopedia of genes and genomes.
- Record: found
- Abstract: found
- Article: not found
Complex heatmaps reveal patterns and correlations in multidimensional genomic data.
- Record: found
- Abstract: found
- Article: not found
Toward understanding the origin and evolution of cellular organisms
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.
Similar content283
- Metabolic Bone Disease in the Context of Metastatic Neuroendocrine Tumor: Differentiation from Skeletal Metastasis, the Molecular PET–CT Imaging Features, and Exploring the Possible Etiopathologies Including Parathyroid Adenoma (MEN1) and Paraneoplastic Humoral Hypercalcemia of Malignancy Due to PTHrP HypersecretionAuthors: Rohit Ranade, Sandip Basu