- Record: found
- Abstract: found
- Article: found
IMPATIENT-qPCR: monitoring SELEX success during in vitro aptamer evolution

Read this article at
Abstract
Abstract
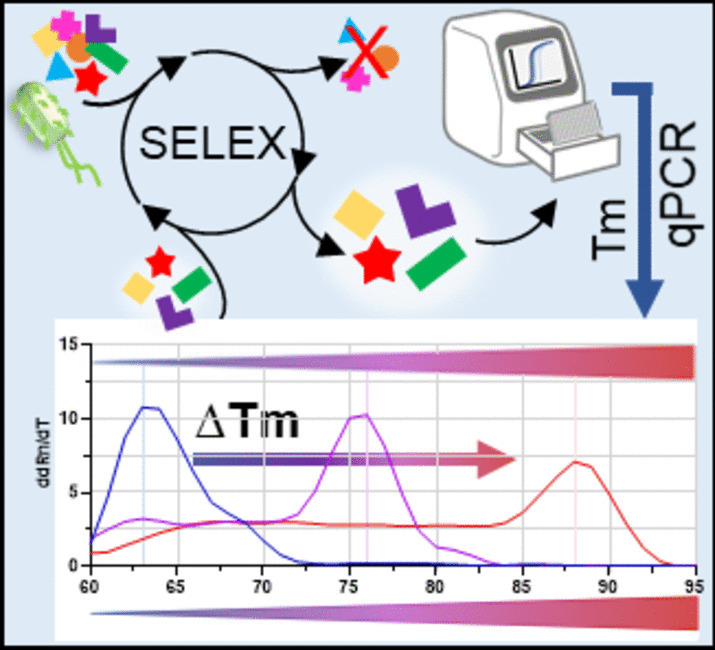
SELEX (Systematic Evolution of Ligands by Exponential enrichment) processes aim on the evolution of high-affinity aptamers as binding entities in diagnostics and biosensing. Aptamers can represent game-changers as constituents of diagnostic assays for the management of instantly occurring infectious diseases or other health threats. Without in-process quality control measures SELEX suffers from low overall success rates. We present a quantitative PCR method for fast and easy quantification of aptamers bound to their targets. Simultaneous determination of melting temperatures ( T m) of each SELEX round delivers information on the evolutionary success via the correlation of increasing GC content and T m alone with a round-wise increase of aptamer affinity to the respective target. Based on nine successful and published previous SELEX processes, in which the evolution/selection of aptamer affinity/specificity was demonstrated, we here show the functionality of the IMPATIENT-qPCR for polyclonal aptamer libraries and resulting individual aptamers. Based on the ease of this new evolution quality control, we hope to introduce it as a valuable tool to accelerate SELEX processes in general.
IMPATIENT-qPCR SELEX success monitoring. Selection and evolution of high-affinity aptamers using SELEX technology with direct aptamer evolution monitoring using melting curve shifting analyses to higher T m by quantitative PCR with fluorescence dye SYBR Green I.
Related collections
Most cited references30
- Record: found
- Abstract: not found
- Article: not found
Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase
- Record: found
- Abstract: found
- Article: not found
In vitro selection of RNA molecules that bind specific ligands.
- Record: found
- Abstract: found
- Article: not found