- Record: found
- Abstract: found
- Article: found
An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction
Read this article at
Abstract
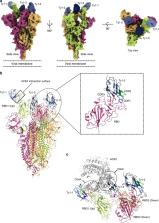
SARS-CoV-2 enters host cells through an interaction between the spike glycoprotein and the angiotensin converting enzyme 2 (ACE2) receptor. Directly preventing this interaction presents an attractive possibility for suppressing SARS-CoV-2 replication. Here, we report the isolation and characterization of an alpaca-derived single domain antibody fragment, Ty1, that specifically targets the receptor binding domain (RBD) of the SARS-CoV-2 spike, directly preventing ACE2 engagement. Ty1 binds the RBD with high affinity, occluding ACE2. A cryo-electron microscopy structure of the bound complex at 2.9 Å resolution reveals that Ty1 binds to an epitope on the RBD accessible in both the ‘up’ and ‘down’ conformations, sterically hindering RBD-ACE2 binding. While fusion to an Fc domain renders Ty1 extremely potent, Ty1 neutralizes SARS-CoV-2 spike pseudovirus as a 12.8 kDa nanobody, which can be expressed in high quantities in bacteria, presenting opportunities for manufacturing at scale. Ty1 is therefore an excellent candidate as an intervention against COVID-19.
Abstract
Here, Hanke et al. immunize an alpaca with SARS-CoV-2 spike protein domains and identify a nanobody that binds the receptor binding domain of spike in both the up and down conformations and sterically hinders ACE2 engagement.
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
- Record: found
- Abstract: found
- Article: not found
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein

- Record: found
- Abstract: found
- Article: found