- Record: found
- Abstract: found
- Article: found
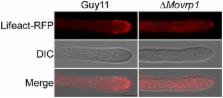
MoVrp1, a putative verprolin protein, is required for asexual development and infection in the rice blast fungus Magnaporthe oryzae
Read this article at
Abstract
Endocytosis is a crucial cellular process in eukaryotic cells which involves clathrin and/or adaptor proteins, lipid kinases, phosphatases and the actin cytoskeleton. Verprolin proteins, such as Vrp1 in Saccharomyces cerevisiae, are conserved family proteins that regulate actin binding and endocytosis. Here, we identified and characterized MoVrp1 as the yeast Vrp1 homolog in Magnaporthe oryzae. Deletion of the MoVRP1 gene resulted in defects in vegetative growth, asexual development, and infection of the host plant. The ∆Movrp1 mutants also exhibited decreased extracellular peroxidase and laccase activities and showed defects in colony pigmentation, hyphal surface hydrophobicity, cell wall integrity, autophagy, endocytosis, and secretion of avirulent effector. Our studies provided new evidences that MoVrp1 involved in actin cytoskeleton is important for growth, morphogenesis, cellular trafficking, and fungal pathogenesis.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast
- Record: found
- Abstract: found
- Article: not found
Metabolism and functions of gamma-aminobutyric acid.
- Record: found
- Abstract: found
- Article: not found