- Record: found
- Abstract: found
- Article: found
Sialidase Inhibitors with Different Mechanisms
Read this article at
Abstract

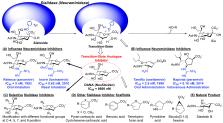
Sialidases, or neuraminidases, are enzymes that catalyze the hydrolysis of sialic acid (Sia)-containing molecules, mostly removal of the terminal Sia (desialylation). By desialylation, sialidase can modulate the functionality of the target compound and is thus often involved in biological pathways. Inhibition of sialidases with inhibitors is an important approach for understanding sialidase function and the underlying mechanisms and could serve as a therapeutic approach as well. Transition-state analogues, such as anti-influenza drugs oseltamivir and zanamivir, are major sialidase inhibitors. In addition, difluoro-sialic acids were developed as mechanism-based sialidase inhibitors. Further, fluorinated quinone methide-based suicide substrates were reported. Sialidase product analogue inhibitors were also explored. Finally, natural products have shown competitive inhibiton against viral, bacterial, and human sialidases. This Perspective describes sialidase inhibitors with different mechanisms and their activities and future potential, which include transition-state analogue inhibitors, mechanism-based inhibitors, suicide substrate inhibitors, product analogue inhibitors, and natural product inhibitors.
Related collections
Most cited references242
- Record: found
- Abstract: found
- Article: not found
New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays.
- Record: found
- Abstract: found
- Article: not found
Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium.
- Record: found
- Abstract: found
- Article: not found