- Record: found
- Abstract: found
- Article: found
Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
Read this article at
Abstract
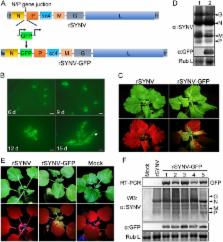
Reverse genetics systems have been established for all major groups of plant DNA and positive-strand RNA viruses, and our understanding of their infection cycles and pathogenesis has benefitted enormously from use of these approaches. However, technical difficulties have heretofore hampered applications of reverse genetics to plant negative-strand RNA (NSR) viruses. Here, we report recovery of infectious virus from cloned cDNAs of a model plant NSR, Sonchus yellow net rhabdovirus (SYNV). The procedure involves Agrobacterium-mediated transcription of full-length SYNV antigenomic RNA and co-expression of the nucleoprotein (N), phosphoprotein (P), large polymerase core proteins and viral suppressors of RNA silencing in Nicotiana benthamiana plants. Optimization of core protein expression resulted in up to 26% recombinant SYNV (rSYNV) infections of agroinfiltrated plants. A reporter virus, rSYNV-GFP, engineered by inserting a green fluorescence protein (GFP) gene between the N and P genes was able to express GFP during systemic infections and after repeated plant-to-plant mechanical passages. Deletion analyses with rSYNV-GFP demonstrated that SYNV cell-to-cell movement requires the sc4 protein and suggested that uncoiled nucleocapsids are infectious movement entities. Deletion analyses also showed that the glycoprotein is not required for systemic infection, although the glycoprotein mutant was defective in virion morphogenesis. Taken together, we have developed a robust reverse genetics system for SYNV that provides key insights into morphogenesis and movement of an enveloped plant virus. Our study also provides a template for developing analogous systems for reverse genetic analysis of other plant NSR viruses.
Author Summary
Reverse genetics is a powerful tool for fundamental studies of virus biology, pathology and biotechnology applications. Although plant negative-strand RNA (NSR) viruses consist of members in the Rhabdoviridae, Bunyaviridae, Ophioviridae families and several unassigned genera that collectively account for many economically important crop diseases, unfortunately, several technical difficulties have hindered application of genetic engineering to these groups of viruses. This study describes the first reverse genetics system developed for plant NSR viruses. We report an efficient procedure for production of infectious virus from cloned cDNAs of sonchus yellow net virus (SYNV) RNAs, a model plant rhabdovirus. We have also engineered a recombinant SYNV vector for stable expression of a fluorescent reporter gene. Using this system, we have generated targeted SYNV mutants whose analyses provide key insights into enveloped plant virus movement and morphogenesis processes. Moreover, our findings provide a template for reverse genetics studies with other plant rhabdoviruses, and a strategy to circumvent technical difficulties that have hampered these applications to plant NSR viruses.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus.
- Record: found
- Abstract: found
- Article: not found