- Record: found
- Abstract: found
- Article: not found
Individual brain organoids reproducibly form cell diversity of the human cerebral cortex
Read this article at
Summary Paragraph
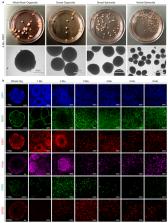
Experimental models of the human brain are needed for basic understanding of its development and disease 1 . Human brain organoids hold unprecedented promise for this purpose; however, they are plagued by high organoid-to-organoid variability 2, 3 . This has raised doubts as to whether developmental processes of the human brain can occur outside the context of embryogenesis with a degree of reproducibility comparable to the endogenous tissue. Here, we show that an organoid model of the dorsal forebrain can achieve reproducible generation of a rich diversity of cell types appropriate for the human cerebral cortex. Using single-cell RNA sequencing of 166,242 cells isolated from 21 individual organoids, we find that 95% of the organoids generate a virtually indistinguishable compendium of cell types, through the same developmental trajectories, and with organoid-to-organoid variability comparable to that of individual endogenous brains. Furthermore, organoids derived from different stem cell lines show consistent reproducibility in the cell types produced. The data demonstrate that reproducible development of complex central nervous system cellular diversity does not require the context of the embryo, and that establishment of terminal cell identity is a highly constrained process that can emerge from diverse stem cell origins and growth environments.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo.
- Record: found
- Abstract: found
- Article: not found
Molecular identity of human outer radial glia during cortical development.
- Record: found
- Abstract: found
- Article: not found
