- Record: found
- Abstract: found
- Article: not found
Screening and druggability analysis of some plant metabolites against SARS-CoV-2: An integrative computational approach
Read this article at
Abstract
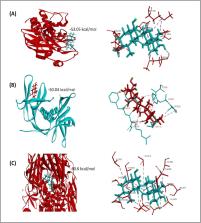
The sudden outbreak of novel coronavirus has caused a global concern due to its infection rate and mortality. Despite extensive research, there are still no specific drugs or vaccines to combat SARS-CoV-2 infection. Hence, this study was designed to evaluate some plant-based active compounds for drug candidacy against SARS-CoV-2 by using virtual screening methods and various computational analyses. A total of 27 plant metabolites were screened against SARS-CoV-2 main protease proteins (MPP), Nsp9 RNA binding protein, spike receptor binding domain, spike ecto-domain and HR2 domain using a molecular docking approach. Four metabolites, i.e., asiatic acid, avicularin, guajaverin, and withaferin showed maximum binding affinity with all key proteins in terms of lowest global binding energy. The crucial binding sites and drug surface hotspots were unravelled for each viral protein. The top candidates were further employed for ADME (absorption, distribution, metabolism, and excretion) analysis to investigate their drug profiles. Results suggest that none of the compounds render any undesirable consequences that could reduce their drug likeness properties. The analysis of toxicity pattern revealed no significant tumorigenic, mutagenic, irritating, or reproductive effects by the compounds. However, withaferin was comparatively toxic among the top four candidates with considerable cytotoxicity and immunotoxicity. Most of the target class by top drug candidates belonged to enzyme groups (e.g. oxidoreductases hydrolases, phosphatases). Moreover, results of drug similarity prediction revealed two approved structural analogs of Asiatic acid i.e. Hydrocortisone (DB00741) (previously used for SARS-CoV-1 and MERS) and Dinoprost-tromethamine (DB01160) from DrugBank. In addition, two other biologically active compounds, Mupirocin (DB00410) and Simvastatin (DB00641) could be an option for the treatment of viral infections. The study may pave the way to develop effective medications and preventive measure against SARS-CoV-2. Due to the encouraging results, we highly recommend further in vivo trials for the experimental validation of our findings.
Graphical abstract
Highlights
-
•
Asiatic acid, Avicularin, Guajaverin & Withaferin showed maximum binding affinity.
-
•
The crucial binding sites & drug surface hotspots were unravelled for key viral protein.
-
•
ADME analysis revealed that top candidates do not show any undesirable consequences.
-
•
Witheferin was comparatively toxic with considerable cytotoxicity and immunotoxicity.
-
•
Hydrocortisone and Simvastatin could be an option to treat SARS-CoV-2 infections.
Related collections
Most cited references99

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients

- Record: found
- Abstract: found
- Article: found

