- Record: found
- Abstract: found
- Article: found
Characterization of Phage Resistance and Their Impacts on Bacterial Fitness in Pseudomonas aeruginosa
Read this article at
ABSTRACT
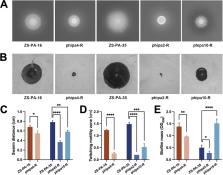
The emergence and spread of antibiotic resistance pose serious environmental and health challenges. Attention has been drawn to phage therapy as an alternative approach to combat antibiotic resistance with immense potential. However, one of the obstacles to phage therapy is phage resistance, and it can be acquired through genetic mutations, followed by consequences of phenotypic variations. Therefore, understanding the mechanisms underlying phage-host interactions will provide us with greater detail on how to optimize phage therapy. In this study, three lytic phages (phipa2, phipa4, and phipa10) were isolated to investigate phage resistance and the potential fitness trade-offs in Pseudomonas aeruginosa. Specifically, in phage-resistant mutants phipa2-R and phipa4-R, mutations in conferring resistance occurred in genes pilT and pilB, both essential for type IV pili (T4P) biosynthesis. In the phage-resistant mutant phipa10-R, a large chromosomal deletion of ~294 kb, including the hmgA (homogentisate 1,2-dioxygenase) and galU (UTP–glucose-1-phosphate uridylyltransferase) genes, was observed and conferred phage phipa10 resistance. Further, we show examples of associated trade-offs in these phage-resistant mutations, e.g., impaired motility, reduced biofilm formation, and increased antibiotic susceptibility. Collectively, our study sheds light on resistance-mediated genetic mutations and their pleiotropic phenotypes, further emphasizing the impressive complexity and diversity of phage-host interactions and the challenges they pose when controlling bacterial diseases in this important pathogen.
IMPORTANCE Battling phage resistance is one of the main challenges faced by phage therapy. To overcome this challenge, detailed information about the mechanisms of phage-host interactions is required to understand the bacterial evolutionary processes. In this study, we identified mutations in key steps of type IV pili (T4P) and O-antigen biosynthesis leading to phage resistance and provided new evidence on how phage predation contributed toward host phenotypes and fitness variations. Together, our results add further fundamental knowledge on phage-host interactions and how they regulate different aspects of Pseudomonas cell behaviors.
Related collections
Most cited references72

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
BEDTools: a flexible suite of utilities for comparing genomic features
- Record: found
- Abstract: found
- Article: not found