- Record: found
- Abstract: found
- Article: found
Dynamical control enables the formation of demixed biomolecular condensates
Read this article at
Abstract
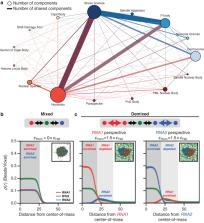
Cellular matter can be organized into compositionally distinct biomolecular condensates. For example, in Ashbya gossypii, the RNA-binding protein Whi3 forms distinct condensates with different RNA molecules. Using criteria derived from a physical framework for explaining how compositionally distinct condensates can form spontaneously via thermodynamic considerations, we find that condensates in vitro form mainly via heterotypic interactions in binary mixtures of Whi3 and RNA. However, within these condensates, RNA molecules become dynamically arrested. As a result, in ternary systems, simultaneous additions of Whi3 and pairs of distinct RNA molecules lead to well-mixed condensates, whereas delayed addition of an RNA component results in compositional distinctness. Therefore, compositional identities of condensates can be achieved via dynamical control, being driven, at least partially, by the dynamical arrest of RNA molecules. Finally, we show that synchronizing the production of different RNAs leads to more well-mixed, as opposed to compositionally distinct condensates in vivo.
Abstract
In this work, the authors report that protein-RNA condensates with shared proteins and distinct RNAs can form and persist in vitro and in cells as distinct entities if the nonshared RNA molecules are dynamically arrested, but the shared protein components are dynamically exchangeable.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.

- Record: found
- Abstract: not found
- Article: not found