- Record: found
- Abstract: found
- Article: found
Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha

Read this article at
Abstract
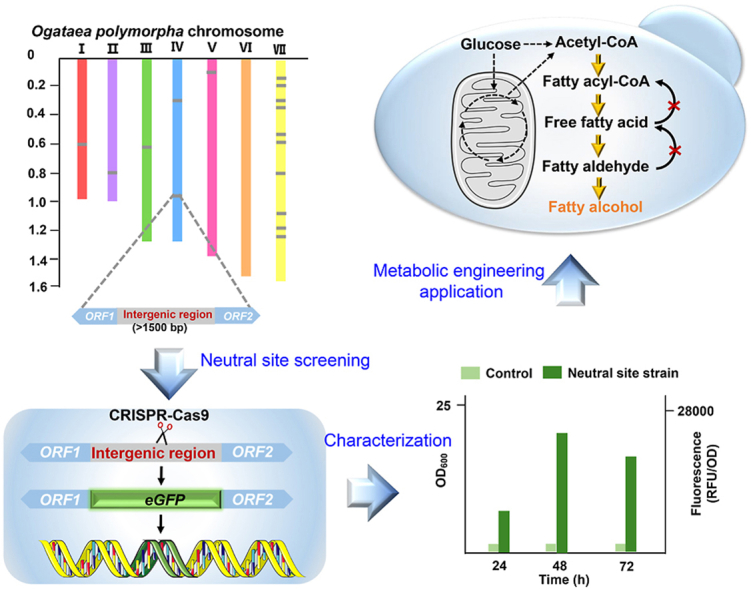
Methylotrophic yeast Ogataea polymorpha is capable to utilize multiple carbon feedstocks especially methanol as sole carbon source and energy, making it an ideal host for bio-manufacturing. However, the lack of gene integration sites limits its systems metabolic engineering, in particular construction of genome-integrated pathway. We here screened the genomic neutral sites for gene integration without affecting cellular fitness, by genomic integration of an enhanced green fluorescent protein ( eGFP) gene via CRISPR-Cas9 technique. After profiling the growth and fluorescent intensity in various media, 17 genome positions were finally identified as potential neutral sites. Finally, integration of fatty alcohol synthetic pathway genes into neutral sites NS2 and NS3, enabled the production of 4.5 mg/L fatty alcohols, indicating that these neutral sites can be used for streamline metabolic engineering in O. polymorpha. We can anticipate that the neutral sites screening method described here can be easily adopted to other eukaryotes.
Graphical abstract
Related collections
Most cited references31

- Record: found
- Abstract: found
- Article: found
CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing
- Record: found
- Abstract: found
- Article: not found
Extraction of genomic DNA from yeasts for PCR-based applications.
- Record: found
- Abstract: not found
- Article: not found