- Record: found
- Abstract: found
- Article: found
New Insights into Somatic Embryogenesis: LEAFY COTYLEDON1, BABY BOOM1 and WUSCHEL-RELATED HOMEOBOX4 Are Epigenetically Regulated in Coffea canephora
Read this article at
Abstract
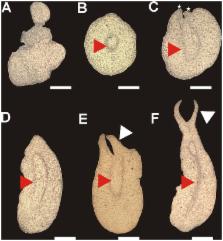
Plant cells have the capacity to generate a new plant without egg fertilization by a process known as somatic embryogenesis (SE), in which differentiated somatic cells can form somatic embryos able to generate a functional plant. Although there have been advances in understanding the genetic basis of SE, the epigenetic mechanism that regulates this process is still unknown. Here, we show that the embryogenic development of Coffea canephora proceeds through a crosstalk between DNA methylation and histone modifications during the earliest embryogenic stages of SE. We found that low levels of DNA methylation, histone H3 lysine 9 dimethylation (H3K9me2) and H3K27me3 change according to embryo development. Moreover, the expression of LEAFY COTYLEDON1 ( LEC1) and BABY BOOM1 ( BBM1) are only observed after SE induction, whereas WUSCHEL-RELATED HOMEOBOX4 ( WOX4) decreases its expression during embryo maturation. Using a pharmacological approach, it was found that 5-Azacytidine strongly inhibits the embryogenic response by decreasing both DNA methylation and gene expression of LEC1 and BBM1. Therefore, in order to know whether these genes were epigenetically regulated, we used Chromatin Immunoprecipitation (ChIP) assays. It was found that WOX4 is regulated by the repressive mark H3K9me2, while LEC1 and BBM1 are epigenetically regulated by H3K27me3. We conclude that epigenetic regulation plays an important role during somatic embryogenic development, and a molecular mechanism for SE is proposed.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana.
- Record: found
- Abstract: found
- Article: not found
Histone methylation in higher plants.
- Record: found
- Abstract: found
- Article: not found