- Record: found
- Abstract: found
- Article: found
The CIAMIB: a Large and Metabolically Diverse Collection of Inflammation-Associated Bacteria from the Murine Gut
Read this article at
ABSTRACT
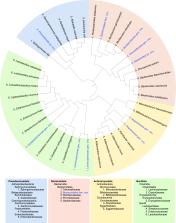
Gut inflammation directly impacts the growth and stability of commensal gut microbes and can lead to long-lasting changes in microbiota composition that can prolong or exacerbate disease states. While mouse models are used extensively to investigate the interplay between microbes and the inflamed state, the paucity of cultured mouse gut microbes has hindered efforts to determine causal relationships. To address this issue, we are assembling the Collection of Inflammation-Associated Mouse Intestinal Bacteria (CIAMIB). The initial release of this collection comprises 41 isolates of 39 unique bacterial species, covering 4 phyla and containing 10 previously uncultivated isolates, including 1 novel family and 7 novel genera. The collection significantly expands the number of available Muribaculaceae, Lachnospiraceae, and Coriobacteriaceae isolates and includes microbes from genera associated with inflammation, such as Prevotella and Klebsiella. We characterized the growth of CIAMIB isolates across a diverse range of nutritional conditions and predicted their metabolic potential and anaerobic fermentation capacity based on the genomes of these isolates. We also provide the first metabolic analysis of species within the genus Adlercreutzia, revealing these representatives to be nitrate-reducing and severely restricted in their ability to grow on carbohydrates. CIAMIB isolates are fully sequenced and available to the scientific community as a powerful tool to study host-microbiota interactions.
Related collections
Most cited references98
- Record: found
- Abstract: found
- Article: not found
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.
- Record: found
- Abstract: found
- Article: not found
SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.
- Record: found
- Abstract: not found
- Article: not found