- Record: found
- Abstract: found
- Article: found
Microbiome–metabolome reveals the contribution of gut–kidney axis on kidney disease
Read this article at
Abstract
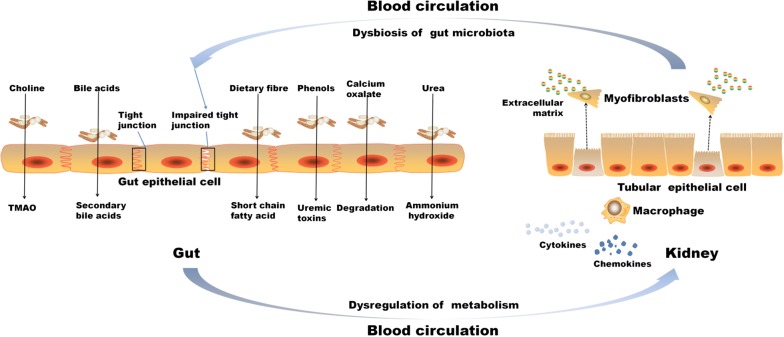
Dysbiosis represents changes in composition and structure of the gut microbiome community (microbiome), which may dictate the physiological phenotype (health or disease). Recent technological advances and efforts in metagenomic and metabolomic analyses have led to a dramatical growth in our understanding of microbiome, but still, the mechanisms underlying gut microbiome–host interactions in healthy or diseased state remain elusive and their elucidation is in infancy. Disruption of the normal gut microbiota may lead to intestinal dysbiosis, intestinal barrier dysfunction, and bacterial translocation. Excessive uremic toxins are produced as a result of gut microbiota alteration, including indoxyl sulphate, p-cresyl sulphate, and trimethylamine-N-oxide, all implicated in the variant processes of kidney diseases development. This review focuses on the pathogenic association between gut microbiota and kidney diseases (the gut–kidney axis), covering CKD, IgA nephropathy, nephrolithiasis, hypertension, acute kidney injury, hemodialysis and peritoneal dialysis in clinic. Targeted interventions including probiotic, prebiotic and symbiotic measures are discussed for their potential of re-establishing symbiosis, and more effective strategies for the treatment of kidney diseases patients are suggested. The novel insights into the dysbiosis of the gut microbiota in kidney diseases are helpful to develop novel therapeutic strategies for preventing or attenuating kidney diseases and complications.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: found
The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data
- Record: found
- Abstract: found
- Article: found
Expansion of Urease- and Uricase-Containing, Indole- and p-Cresol-Forming and Contraction of Short-Chain Fatty Acid-Producing Intestinal Microbiota in ESRD
- Record: found
- Abstract: found
- Article: not found