- Record: found
- Abstract: found
- Article: found
Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance
Read this article at
Abstract
Background
Recently, we reported that some dairy cows could produce high amounts of milk with high amounts of protein (defined as milk protein yield [MPY]) when a population was raised under the same nutritional and management condition, a potential new trait that can be used to increase high-quality milk production. It is unknown to what extent the rumen microbiome and its metabolites, as well as the host metabolism, contribute to MPY. Here, analysis of rumen metagenomics and metabolomics, together with serum metabolomics was performed to identify potential regulatory mechanisms of MPY at both the rumen microbiome and host levels.
Results
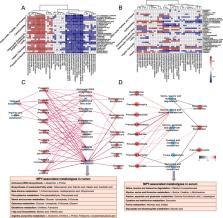
Metagenomics analysis revealed that several Prevotella species were significantly more abundant in the rumen of high-MPY cows, contributing to improved functions related to branched-chain amino acid biosynthesis. In addition, the rumen microbiome of high-MPY cows had lower relative abundances of organisms with methanogen and methanogenesis functions, suggesting that these cows may produce less methane. Metabolomics analysis revealed that the relative concentrations of rumen microbial metabolites (mainly amino acids, carboxylic acids, and fatty acids) and the absolute concentrations of volatile fatty acids were higher in the high-MPY cows. By associating the rumen microbiome with the rumen metabolome, we found that specific microbial taxa (mainly Prevotella species) were positively correlated with ruminal microbial metabolites, including the amino acids and carbohydrates involved in glutathione, phenylalanine, starch, sucrose, and galactose metabolism. To detect the interactions between the rumen microbiome and host metabolism, we associated the rumen microbiome with the host serum metabolome and found that Prevotella species may affect the host’s metabolism of amino acids (including glycine, serine, threonine, alanine, aspartate, glutamate, cysteine, and methionine). Further analysis using the linear mixed effect model estimated contributions to the variation in MPY based on different omics and revealed that the rumen microbial composition, functions, and metabolites, and the serum metabolites contributed 17.81, 21.56, 29.76, and 26.78%, respectively, to the host MPY.
Conclusions
These findings provide a fundamental understanding of how the microbiome-dependent and host-dependent mechanisms contribute to varied individualized performance in the milk production quality of dairy cows under the same management condition. This fundamental information is vital for the development of potential manipulation strategies to improve milk quality and production through precision feeding.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
MetPA: a web-based metabolomics tool for pathway analysis and visualization.
- Record: found
- Abstract: found
- Article: not found
