- Record: found
- Abstract: found
- Article: found
Global Analysis of Photosynthesis Transcriptional Regulatory Networks
Read this article at
Abstract
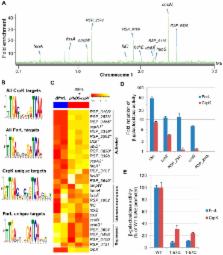
Photosynthesis is a crucial biological process that depends on the interplay of many components. This work analyzed the gene targets for 4 transcription factors: FnrL, PrrA, CrpK and MppG (RSP_2888), which are known or predicted to control photosynthesis in Rhodobacter sphaeroides. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) identified 52 operons under direct control of FnrL, illustrating its regulatory role in photosynthesis, iron homeostasis, nitrogen metabolism and regulation of sRNA synthesis. Using global gene expression analysis combined with ChIP-seq, we mapped the regulons of PrrA, CrpK and MppG. PrrA regulates ∼34 operons encoding mainly photosynthesis and electron transport functions, while CrpK, a previously uncharacterized Crp-family protein, regulates genes involved in photosynthesis and maintenance of iron homeostasis. Furthermore, CrpK and FnrL share similar DNA binding determinants, possibly explaining our observation of the ability of CrpK to partially compensate for the growth defects of a ΔFnrL mutant. We show that the Rrf2 family protein, MppG, plays an important role in photopigment biosynthesis, as part of an incoherent feed-forward loop with PrrA. Our results reveal a previously unrealized, high degree of combinatorial regulation of photosynthetic genes and significant cross-talk between their transcriptional regulators, while illustrating previously unidentified links between photosynthesis and the maintenance of iron homeostasis.
Author Summary
Photosynthetic organisms are among the most abundant life forms on earth. Their unique ability to harvest solar energy and use it to fix atmospheric carbon dioxide is at the foundation of the global food chain. This paper reports the first comprehensive analysis of networks that control expression of photosynthesis genes using Rhodobacter sphaeroides, a microbe that has been studied for decades as a model of solar energy capture and other aspects of the photosynthetic lifestyle. We find a previously unappreciated complexity in the level of control of photosynthetic genes, while identifying new links between photosynthesis and central processes like iron availability. This organism is an ancestor of modern day plants, so our data can inform studies in other photosynthetic organisms and improve our ability to harness solar energy for food and industrial processes.
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
The Pfam protein families database.
- Record: found
- Abstract: found
- Article: not found