- Record: found
- Abstract: found
- Article: found
Diffusion tensor imaging and tractwise fractional anisotropy statistics: quantitative analysis in white matter pathology
research-article
Hans-Peter Mueller
1
,
,
Alexander Unrath
1 ,
Anne D Sperfeld
1 ,
Albert C Ludolph
1 ,
Axel Riecker
1 ,
Jan Kassubek
1
9 November 2007
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Background
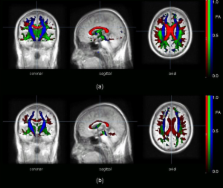
Information on anatomical connectivity in the brain by measurements of the diffusion of water in white matter tracts lead to quantification of local tract directionality and integrity.
Methods
The combination of connectivity mapping (fibre tracking, FT) with quantitative diffusion fractional anisotropy (FA) mapping resulted in the approach of results based on group-averaged data, named tractwise FA statistics (TFAS). The task of this study was to apply these methods to group-averaged data from different subjects to quantify differences between normal subjects and subjects with defined alterations of the corpus callosum (CC).
Related collections
Most cited references27
- Record: found
- Abstract: not found
- Article: not found
Spatial registration and normalization of images
Karl. J. Friston, J. Ashburner, C D Frith … (1995)
- Record: found
- Abstract: not found
- Article: not found
Automatic 3D Intersubject Registration of MR Volumetric Data in Standardized Talairach Space
D. Collins, Peter Neelin, Terrence M. Peters … (1994)
- Record: found
- Abstract: found
- Article: not found
Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space.
D L Collins, P Neelin, T Peters … (1994)